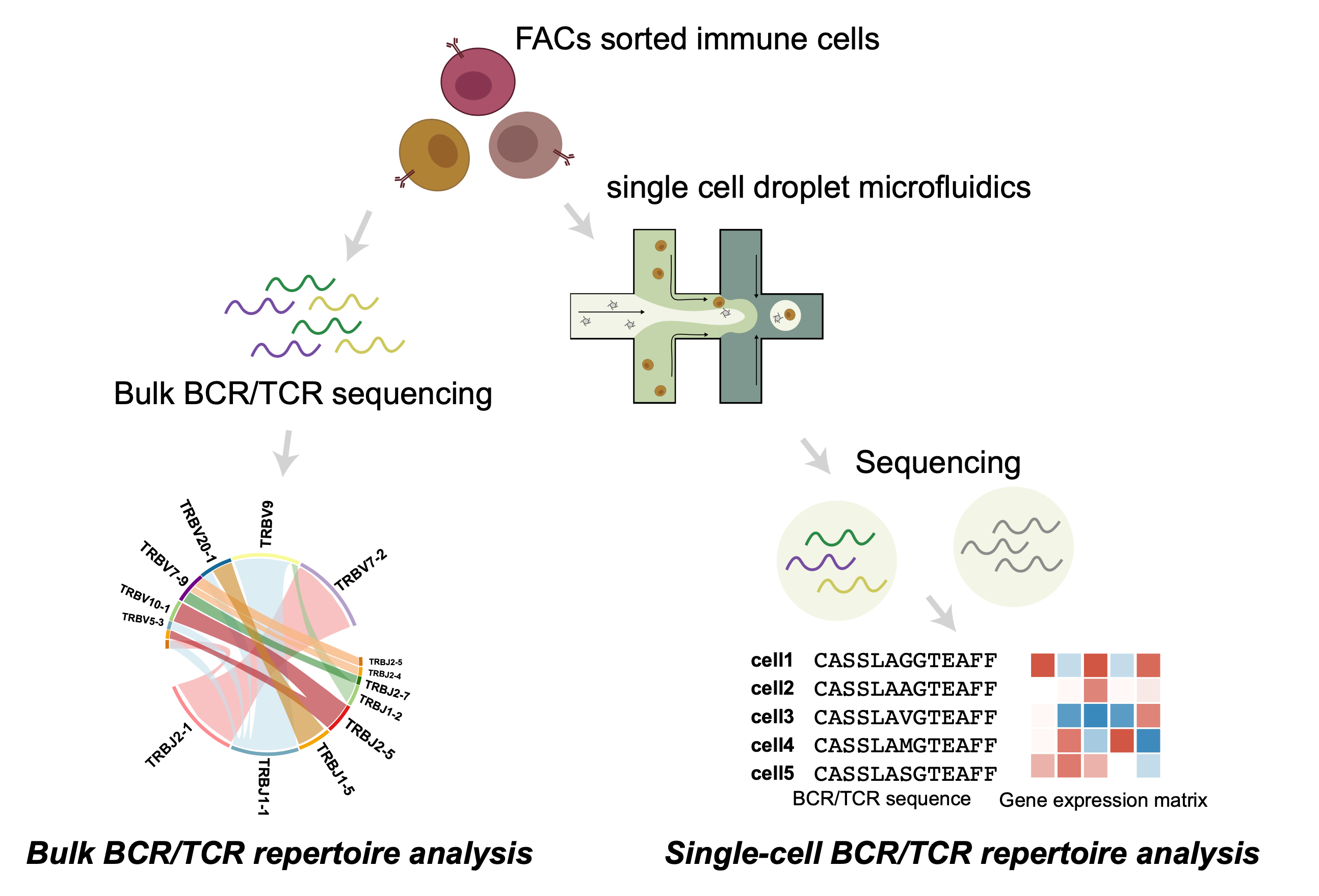
Understand the mystery of antigen receptors
Both B cells and T cells serve a crucial role in the adaptive immune response to antigens. Antibodies secreted by plasma B cells can bind to specific proteins from bacteria and viruses and neutralize these pathogenic antigens. The heavy and light chains of the antibodies, which also compose the B cell receptors (BCRs), have a critical role in recognizing the epitope of the antigens. The antibodies and BCRs are highly diverse, resulting in almost infinite diversity to recognize a great variety of antigens
Like B cells, T cells are produced in the bone marrow, and their surface displays antigen receptors called T cell receptors (TCRs). In the T-cell mediated adaptive immune response, protein antigens are presented to TCRs by major histocompatibility complex (MHC) by antigen-presenting cells (APCs). The binding between TCRs and antigens activates T cells and triggers T cell response.