(Part I) Integration, cross-atlas comparison, and transfer learning
Contents
(Part I) Integration, cross-atlas comparison, and transfer learning¶
This notebook contains codes for integration, cross-atlas comparison, and transfer learning using scAtlasVAE.
Please check the package version at https://github.com/WanluLiuLab/scAtlasVAE/blob/master/environment.yml for reproducing the results.
For more information about the scAtlasVAE model, please see https://scatlasvae.readthedocs.io/en/latest/.
For retrieving datasets, please see https://zenodo.org/records/10472914.
Installing scAtlasVAE
in bash, run pip install scatlasvae
Please run the following code block for importing packages
import scatlasvae
# import packages
import scanpy as sc # import scanpy
import matplotlib
import matplotlib.pyplot as plt # import matplotlib
import numpy as np # import numpy
import pandas as pd # import pandas
import gc # import garbage collector
from typing import Literal, Union # import typing
# set plot linewidth
def setPltLinewidth(linewidth:float): # define function to set plot linewidth
matplotlib.rcParams['axes.linewidth'] = linewidth # set plot linewidth
setPltLinewidth(1) # set plot linewidth to 1
# set plot parameters
plt.rcParams['figure.dpi'] = 300 # s get figure resolution
plt.rcParams['savefig.dpi'] = 300 # set figure resolution
plt.rcParams['font.size'] = 8 # set font size
plt.rcParams['axes.linewidth'] = 1 # set plot linewidth
plt.rcParams['font.family'] = "Arial" # set font family
# Useful functions
try:
import seaborn as sns
import matplotlib
import matplotlib.pyplot as plt
matplotlib.rcParams["font.family"] = "Arial"
matplotlib.rcParams["font.size"] = "10"
matplotlib.rcParams["font.weight"] = 100
matplotlib.rcParams["axes.linewidth"] = 2
matplotlib.rcParams["axes.edgecolor"] = "#000000"
def createFig(figsize=(8, 4)):
fig, ax = plt.subplots()
ax.spines["right"].set_color("none")
ax.spines["top"].set_color("none")
# ax.spines['bottom'].set_color('none')
# ax.spines['left'].set_color('none')
for line in ax.yaxis.get_ticklines():
line.set_markersize(5)
line.set_color("#585958")
line.set_markeredgewidth(0.5)
for line in ax.xaxis.get_ticklines():
line.set_markersize(5)
line.set_markeredgewidth(0.5)
line.set_color("#585958")
ax.set_xbound(0, 10)
ax.set_ybound(0, 10)
fig.set_size_inches(figsize)
return fig, ax
def createSubplots(nrow, ncol, figsize=(8, 8), gridspec_kw={}):
fig, axes = plt.subplots(nrow, ncol, gridspec_kw=gridspec_kw)
for ax in axes.flatten():
ax.spines["right"].set_color("none")
ax.spines["top"].set_color("none")
for line in ax.yaxis.get_ticklines():
line.set_markersize(5)
line.set_color("#585958")
line.set_markeredgewidth(0.5)
for line in ax.xaxis.get_ticklines():
line.set_markersize(5)
line.set_markeredgewidth(0.5)
line.set_color("#585958")
fig.set_size_inches(figsize)
return fig, axes
hist_kws = {"linewidth": 0, "alpha": 0.5}
except:
print("failed to load plotting packages")
from collections import Counter
def plot_a_by_b(adata, a, b):
B = pd.DataFrame(
list(
adata.obs[list(map(lambda x: type(x) == str, adata.obs[a]))]
.groupby(b)
.agg({a: lambda x: dict(Counter(x))})
.iloc[:, 0]
)
).fillna(0)
B = pd.DataFrame((B.to_numpy().T / B.to_numpy().sum(1)).T, columns=B.columns)
B.insert(0, b, pd.Categorical(adata.obs[b]).categories)
ax = B.plot(
x=b,
kind="bar",
stacked=True,
color=sc.pl._tools.scatterplots._get_palette(adata, a),
)
return B, ax
def pandas_aggregation_to_wide(agg_df):
return pd.DataFrame(agg_df.index.tolist(), columns=agg_df.index.names).join(
pd.DataFrame(agg_df.to_numpy(), columns=agg_df.columns)
)
from matplotlib.patches import PathPatch
def adjust_box_widths(g, fac):
"""
Adjust the withs of a seaborn-generated boxplot.
"""
# iterating through Axes instances
for ax in g.axes:
# iterating through axes artists:
for c in ax.get_children():
# searching for PathPatches
if isinstance(c, PathPatch):
# getting current width of box:
p = c.get_path()
verts = p.vertices
verts_sub = verts[:-1]
xmin = np.min(verts_sub[:, 0])
xmax = np.max(verts_sub[:, 0])
xmid = 0.5*(xmin+xmax)
xhalf = 0.5*(xmax - xmin)
# setting new width of box
xmin_new = xmid-fac*xhalf
xmax_new = xmid+fac*xhalf
verts_sub[verts_sub[:, 0] == xmin, 0] = xmin_new
verts_sub[verts_sub[:, 0] == xmax, 0] = xmax_new
# setting new width of median line
for l in ax.lines:
if np.all(l.get_xdata() == [xmin, xmax]):
l.set_xdata([xmin_new, xmax_new])
import scanpy as sc
default_10 = ['#1f77b4',
'#ff7f0e',
'#2ca02c',
'#d62728',
'#9467bd',
'#8c564b',
'#e377c2',
'#7f7f7f',
'#bcbd22',
'#17becf']
zheng_2021_annotation_cmap_cd8 = {
"CD8.c01.Tn.MAL": "#96C3D8",
"CD8.c02.Tm.IL7R": "#5D9BBE",
"CD8.c03.Tm.RPS12": "#F5B375",
"CD8.c04.Tm.CD52": "#C0937E",
"CD8.c05.Tem.CXCR5": "#67A59B",
"CD8.c06.Tem.GZMK": "#A4D38E",
"CD8.c07.Temra.CX3CR1": "#4A9D47",
"CD8.c08.Tk.TYROBP": "#F19294",
"CD8.c09.Tk.KIR2DL4": "#E45A5F",
"CD8.c10.Trm.ZNF683": "#3477A9",
"CD8.c11.Tex.PDCD1": "#BDA7CB",
"CD8.c12.Tex.CXCL13": "#684797",
"CD8.c13.Tex.myl12a": "#9983B7",
"CD8.c14.Tex.TCF7": "#CD9A99",
"CD8.c15.ISG.IFIT1": "#DD4B52",
"CD8.c16.MAIT.SLC4A10": "#DA8F6F",
"CD8.c17.Tm.NME1": "#F58135",
}
zheng_2021_annotation_cmap_cd4 = {
"CD4.c01.Tn.TCF7": "#78AECB",
"CD4.c02.Tn.PASK": "#639FB0",
"CD4.c03.Tn.ADSL": "#98C7A5",
"CD4.c04.Tn.il7r": "#83C180",
"CD4.c05.Tm.TNF": "#B2A4A5",
"CD4.c06.Tm.ANXA1": "#EC8D63",
"CD4.c07.Tm.ANXA2": "#CFC397",
"CD4.c08.Tm.CREM": "#F6B279",
"CD4.c09.Tm.CCL5": "#6197B4",
"CD4.c10.Tm.CAPG": "#CEA168",
"CD4.c11.Tm.GZMA": "#A0A783",
"CD4.c12.Tem.GZMK": "#9ACC90",
"CD4.c13.Temra.CX3CR1": "#6A9A52",
"CD4.c14.Th17.SLC4A10": "#E97679",
"CD4.c15.Th17.IL23R": "#DE4247",
"CD4.c16.Tfh.CXCR5": "#A38CBD",
"CD4.c17.TfhTh1.CXCL13": "#795FA3",
"CD4.c18.Treg.RTKN2": "#E0C880",
"CD4.c19.Treg.S1PR1": "#C28B65",
"CD4.c20.Treg.TNFRSF9": "#A65A34",
"CD4.c21.Treg.OAS1": "#DE4B3F",
"CD4.c22.ISG.IFIT1": "#DD9E82",
"CD4.c23.Mix.NME1": "#E78B75",
"CD4.c24.Mix.NME2": "#F7A96C",
"undefined": "#FFFFFF",
}
zheng_2021_annotation_cmap = zheng_2021_annotation_cmap_cd8.copy()
zheng_2021_annotation_cmap.update(zheng_2021_annotation_cmap_cd4)
chu_annotation_string = """
CD8-3 CD8_c3_Tn #E9ADC2
CD8-13 CD8_c13_Tn_TCF7 #AACC65
CD8-0 CD8_c0_Teff #00AFCA
CD8-2 CD8_c2_Teff #BBB7CB
CD8-8 CD8_c8_Teff_KLRG1 #E1A276
CD8-10 CD8_c10_Teff_CD244 #A5A2B3
CD8-11 CD8_c11_Teff_SEMA4A #A3AFA9
CD8-6 CD8_c6_Tcm #DD7A80
CD8-12 CD8_c12_Trm #A4BD83
CD8-7 CD8_c7_Tpex #EB9B7F
CD8-1 CD8_c1_Tex #76BCD8
CD8-4 CD8_c4_Tstr #E27C97
CD8-5 CD8_c5_Tisg #DF6C87
CD8-9 CD8_c9_Tsen #CCA891
CD4-2 CD4_c2_Tn #E0C8D9
CD4-6 CD4_c6_Tn_FHIT #F0A683
CD4-7 CD4_c7_Tn_TCEA3 #E5AE7C
CD4-9 CD4_c9_Tn_TCF7_SLC40A1 #A6AEBE
CD4-10 CD4_c10_Tn_LEF1_ANKRD55 #B3C28B
CD4-0 CD4_c0_Tcm #4CBBD2
CD4-5 CD4_c5_CTL #E9949E
CD4-1 CD4_c1_Treg #9FC6DB
CD4-3 CD4_c3_TFH #EFB3CC
CD4-8 CD4_c8_Th17 #C4ADA6
CD4-4 CD4_c4_Tstr #EF9AB9
CD4-11 CD4_c11_Tisg #C4D960
"""
subtype_color = {
"Tn": "#CEBF8F",
"Tcm": "#ffbb78",
"Early Tcm/Tem": "#ff7f0e",
"GZMK+ Tem": "#d62728",
"GNLY+ Temra": "#8c564b",
"CMC1+ Temra": "#e377c2",
"ZNF683+ Teff": "#6f3e7c",
"MAIT": "#17becf",
"ILTCK": "#aec7e8",
"ITGAE+ Trm": "#279e68",
"CREM+ Trm": "#aa40fc",
"ITGB2+ Trm": "#5ce041",
"Tpex": "#ff9896",
"GZMK+ Tex": "#C5B0D5",
"ITGAE+ Tex": "#C3823E",
"S100A11+ Tex": "#b5bd61",
"MACF1+ T": "#3288c9",
"Cycling T": "#f7b6d2",
}
subtype_color_alt = {"CD8+ " + k: v for k, v in subtype_color.items()}
chu_annotation = chu_annotation_string.split("\n")[1:-1]
chu_annotation = list(map(lambda x: x.split("\t"), chu_annotation))
import pandas as pd
chu_annotation = pd.DataFrame(chu_annotation)
chu_annotation_name = dict(zip(chu_annotation.iloc[:, 0], chu_annotation.iloc[:, 1]))
chu_annotation_cmap = dict(zip(chu_annotation.iloc[:, 1], chu_annotation.iloc[:, 2]))
chu_annotation_cmap_2 = dict(zip(chu_annotation.iloc[:, 0], chu_annotation.iloc[:, 2]))
default_20 = sc.pl.palettes.default_20
default_28 = sc.pl.palettes.default_28
godsnot_102 = sc.pl.palettes.godsnot_102
Initial integration of pan-disease CD8 atlas¶
adata_cd8 = sc.read_h5ad("./huARdb_v2_GEX.CD8.hvg4k.h5ad")
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8,
batch_key="sample_name",
additional_batch_keys=["study_name"],
batch_embedding="embedding",
batch_hidden_dim=64,
)
vae_model.fit(max_epoch=10, lr=5e-5)
adata_cd8.obsm["X_gex"] = vae_model.get_latent_embedding(show_progress=True)
sc.tl.paga(adata_cd8, groups='cell_subtype_3')
sc.pl.paga(adata_cd8, show=False)
sc.tl.umap(adata_cd8, init_pos='paga')
sc.pp.neighbors(
adata_cd8, use_rep="X_gex", n_neighbors=40
) # compute neighborhood graph
sc.tl.leiden(
adata_cd8, resolution=1.2, key_added="leiden_n_neighbors_40_reoslution_1.2"
) # compute leiden clustering
fig, ax = createFig(figsize=(4, 4))
sc.pl.umap(adata_cd8, color="leiden_n_neighbors_40_reoslution_1.2", ax=ax)
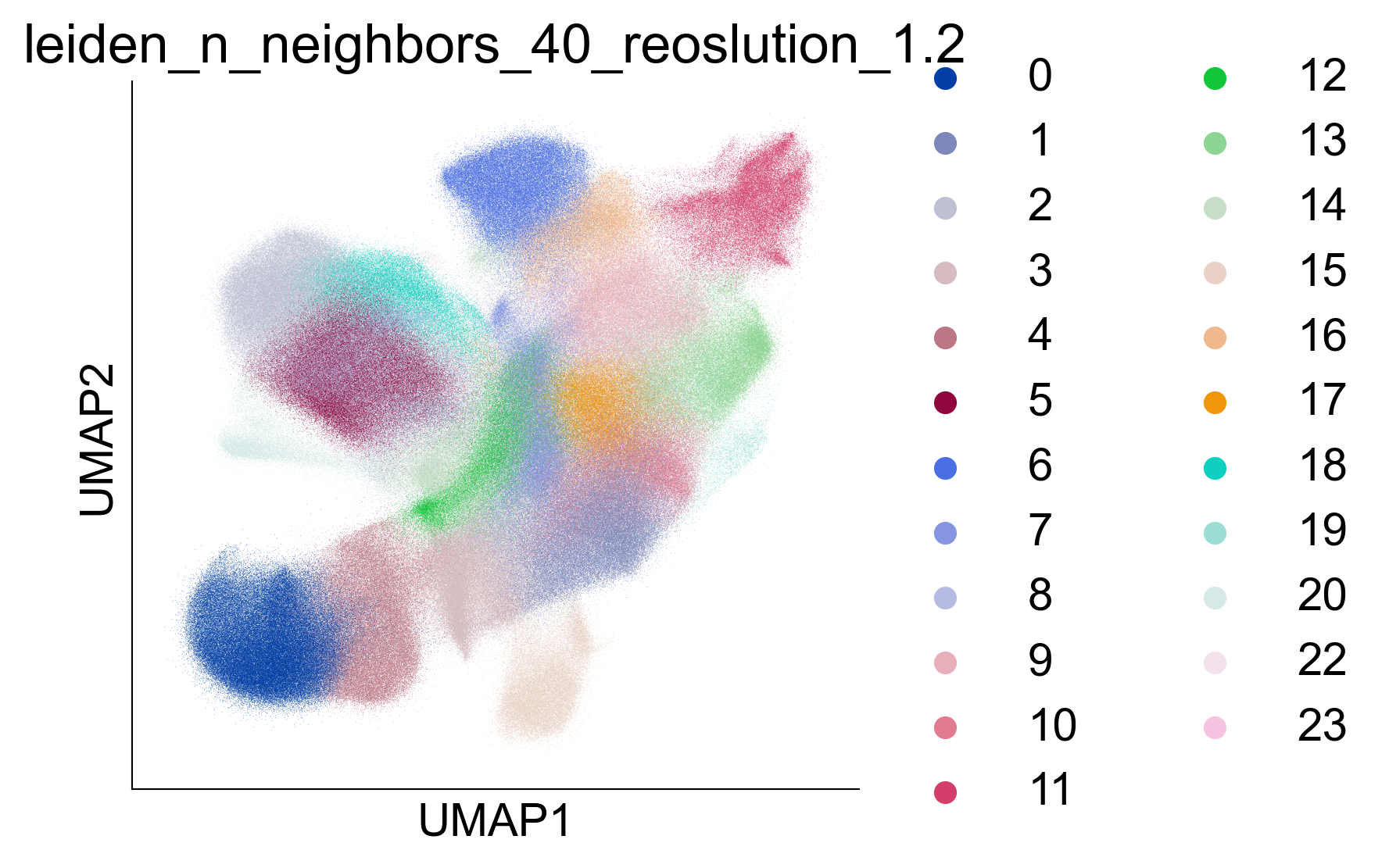
Figure 2A¶
Uniform manifold approximation and projection (UMAP) representation of 1,151,678 cells of the CD8+ T cell atlas, colored by 18 CD8+ T cell subtypes annotated in this study
fig, ax = createFig(figsize=(3, 3))
sc.pl.umap(adata_cd8, color="cell_subtype_3", ax=ax, palette=subtype_color)
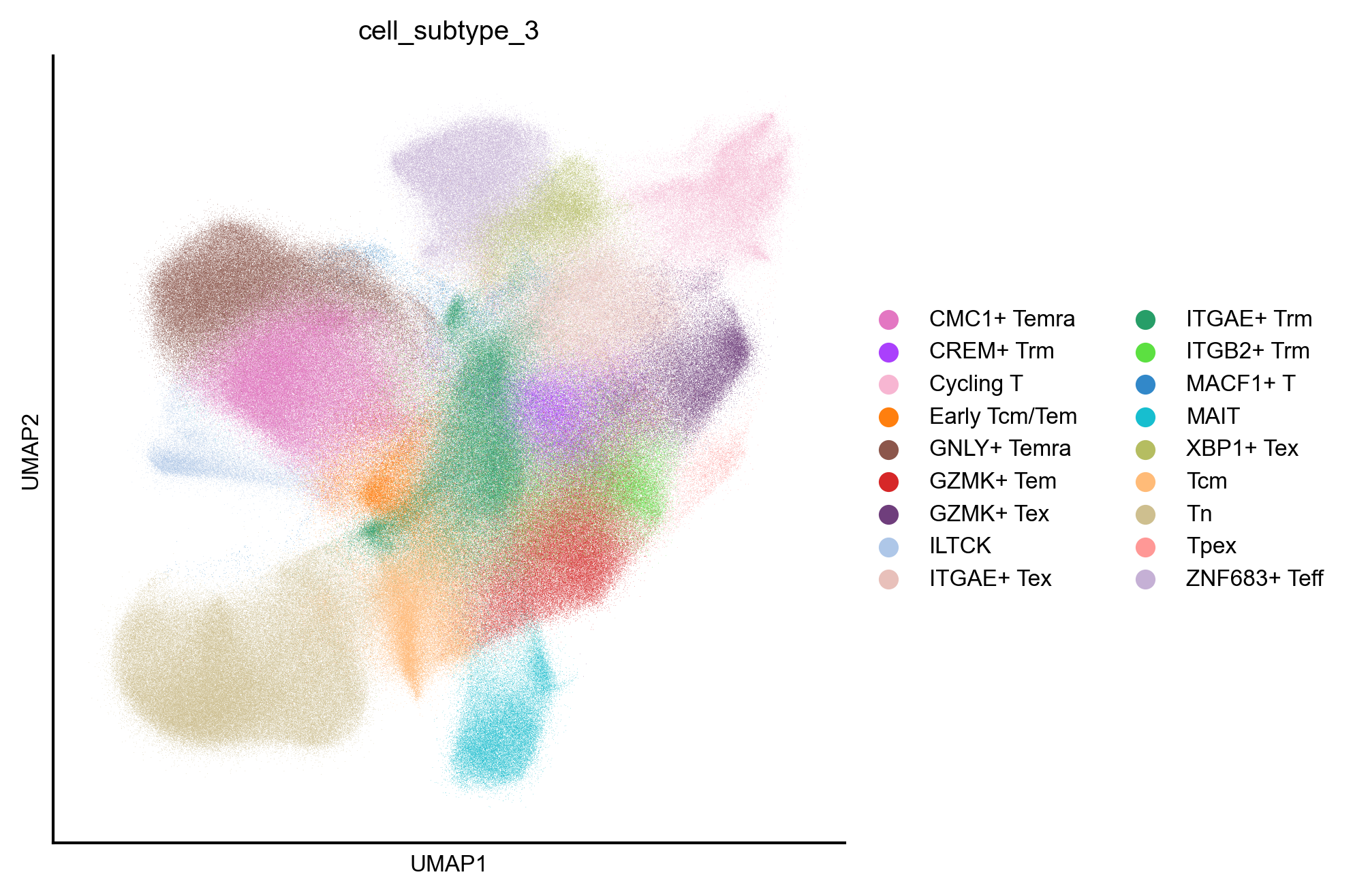
Cross-atlas integration between pan-cancer and pan-disease CD8 atlas¶
adata_cd8_zheng_2021 = sc.read_h5ad("../data/adata_cd8_zheng_2021.h5ad")
adata_cd8_chu_2023 = sc.read_h5ad("../data/adata_cd8_chu_2023.h5ad")
Cross-atlas integration between pan-disease CD8 atlas and Zheng et al., 2021 TCellLandscape¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
adata_cd8_zheng_2021.obs['cell_subtype_3'] = 'undefined'
adata_cd8.obs['cell_subtype_zheng_2021'] = 'undefined'
adata_cd8.obs['atlas_name'] = 'huARdbv2'
adata_cd8_zheng_2021_merged = sc.concat([
adata_cd8_zheng_2021, adata_cd8
], join='inner')
adata_cd8_zheng_2021_merged.obs['cell_subtype_3'] = list(adata_cd8_zheng_2021_merged.obs['cell_subtype_3'])
adata_cd8_zheng_2021_merged.obs['cell_subtype_3'] = adata_cd8_zheng_2021_merged.obs['cell_subtype_3'].fillna("undefined")
multi_atlas_vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8_zheng_2021_merged,
batch_key='sample_name',
additional_batch_keys=['study_name','atlas_name'],
batch_embedding='embedding',
batch_hidden_dim=64,
label_key='cell_subtype_3',
additional_label_keys=['cell_subtype_zheng_2021'],
device='cuda:2'
)
multi_atlas_vae_model.fit(
max_epoch=32,
lr=5e-5,
kl_weight=1.,
n_epochs_kl_warmup=16
)
import umap
adata_cd8_zheng_2021_merged.obsm['X_gex'] = multi_atlas_vae_model.get_latent_embedding()
adata_cd8_zheng_2021_merged.obsm['X_umap'] = umap.UMAP().fit_transform(adata_cd8_zheng_2021_merged.obsm['X_gex'])
predictions = multi_atlas_vae_model.predict_labels(return_pandas=True)
predictions_logits = multi_atlas_vae_model.predict_labels(return_pandas=False)
adata_cd8_zheng_2021_merged.uns['cell_subtype_3_prediction_logits'] = predictions_logits[0].detach().cpu().numpy()
adata_cd8_zheng_2021_merged.uns['cell_subtype_zheng_2021_prediction_logits'] = predictions_logits[1][0].detach().cpu().numpy()
adata_cd8_zheng_2021_merged.obs['cell_subtype_3_prediction'] = list(predictions['cell_subtype_3'])
adata_cd8_zheng_2021_merged.obs['cell_subtype_zheng_2021_prediction'] = list(predictions['cell_subtype_zheng_2021'])
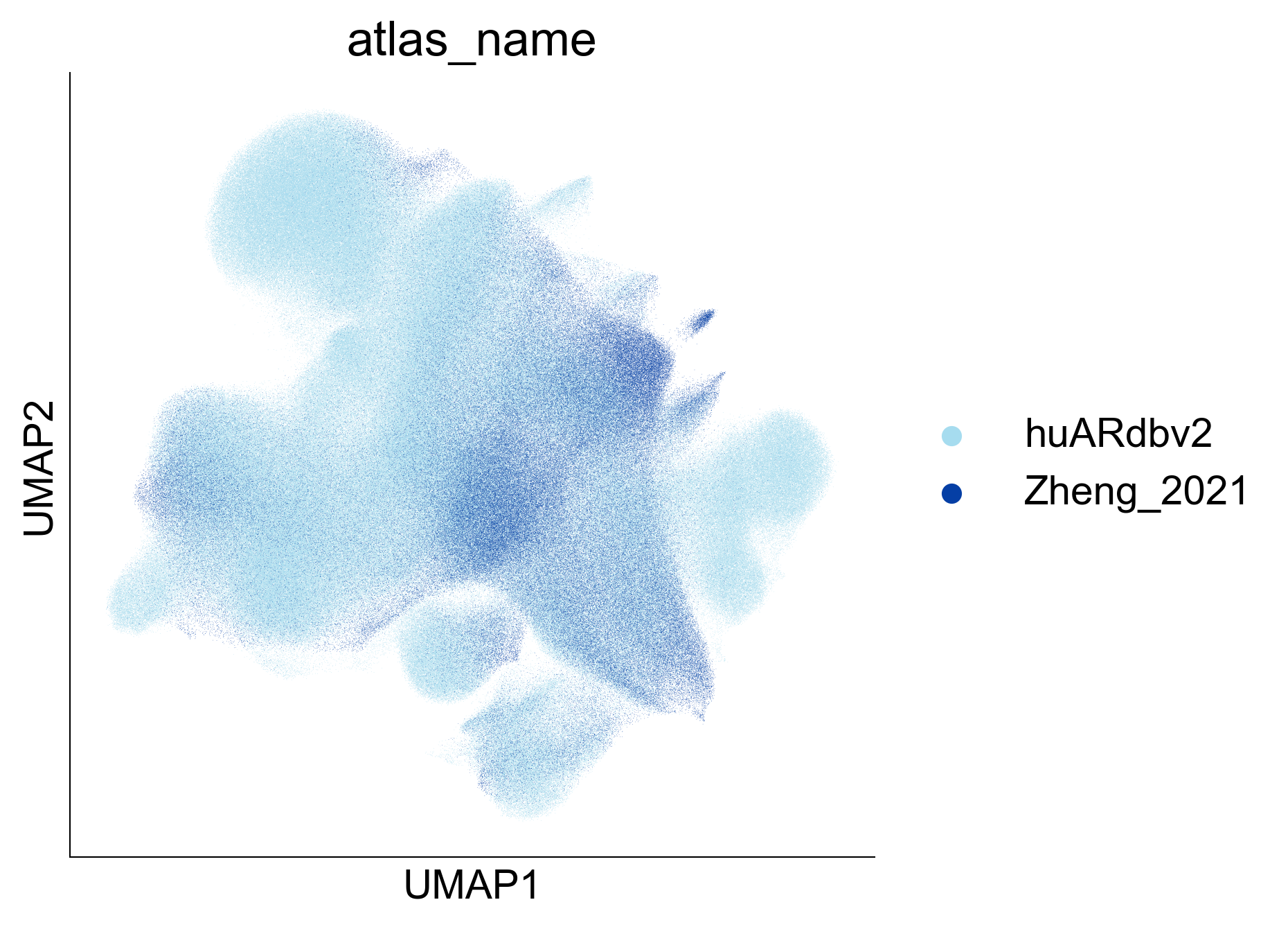
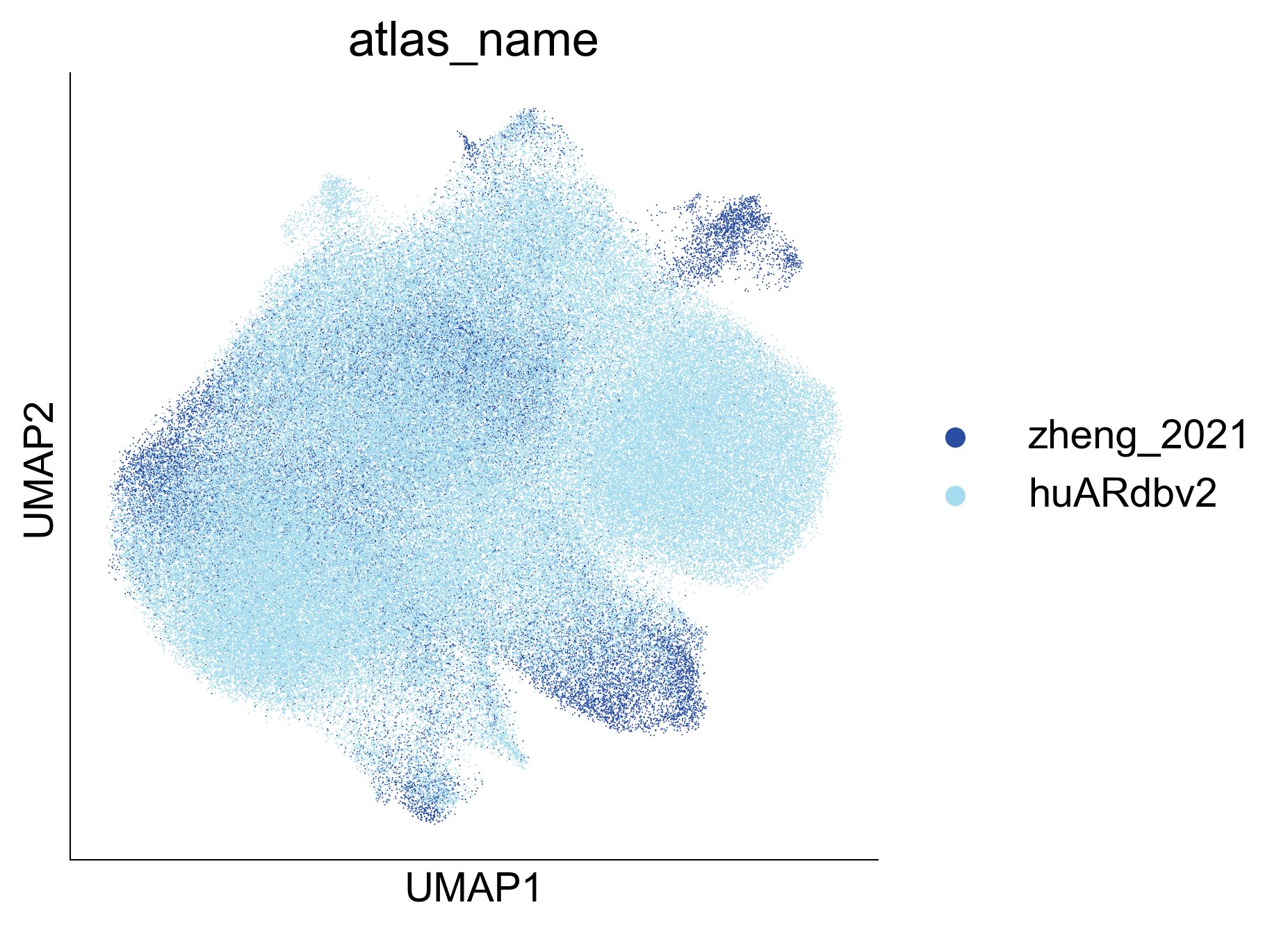
Extended Data Figure 9A¶
fig,ax=createFig(figsize=(5,5))
sc.pl.umap(adata_cd8_zheng_2021_merged[adata_cd8_zheng_2021_merged.obs['atlas_name'] == 'huARdbv2'], color='atlas_name', ax=ax, palette=
{
'Zheng_2021': '#053FA5',
'huARdbv2': '#A6DCEF'
}, show=False, s=0.1
)
sc.pl.umap(adata_cd8_zheng_2021_merged[adata_cd8_zheng_2021_merged.obs['atlas_name'] != 'huARdbv2'], color='atlas_name', ax=ax, palette=
{
'Zheng_2021': '#053FA5',
'huARdbv2': '#A6DCEF'
}, show=False, s=0.1
)
fig.savefig("/Users/snow/Desktop/tmp.png")
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from int32. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from int32. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
zheng_2021_annotation_cmap_cd8['CD8.c11.Tex.PDCD1'] = '#21D5CE'
fig,ax=createSubplots(1,2,figsize=(10,5))
sc.pl.umap(adata_cd8_zheng_2021_merged[
adata_cd8_zheng_2021_merged.obs['zheng_cell_subtype_2021'] != 'undefined'
], color='zheng_cell_subtype_2021', ax=ax[0], show=False, palette=zheng_2021_annotation_cmap_cd8, frameon=False, legend_loc='none')
sc.pl.umap(adata_cd8_zheng_2021_merged[
adata_cd8_zheng_2021_merged.obs['cell_subtype_3'] != 'undefined'
], color='cell_subtype_3', ax=ax[1], palette=subtype_color, frameon=False, legend_loc='none')
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from int32. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from int32. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
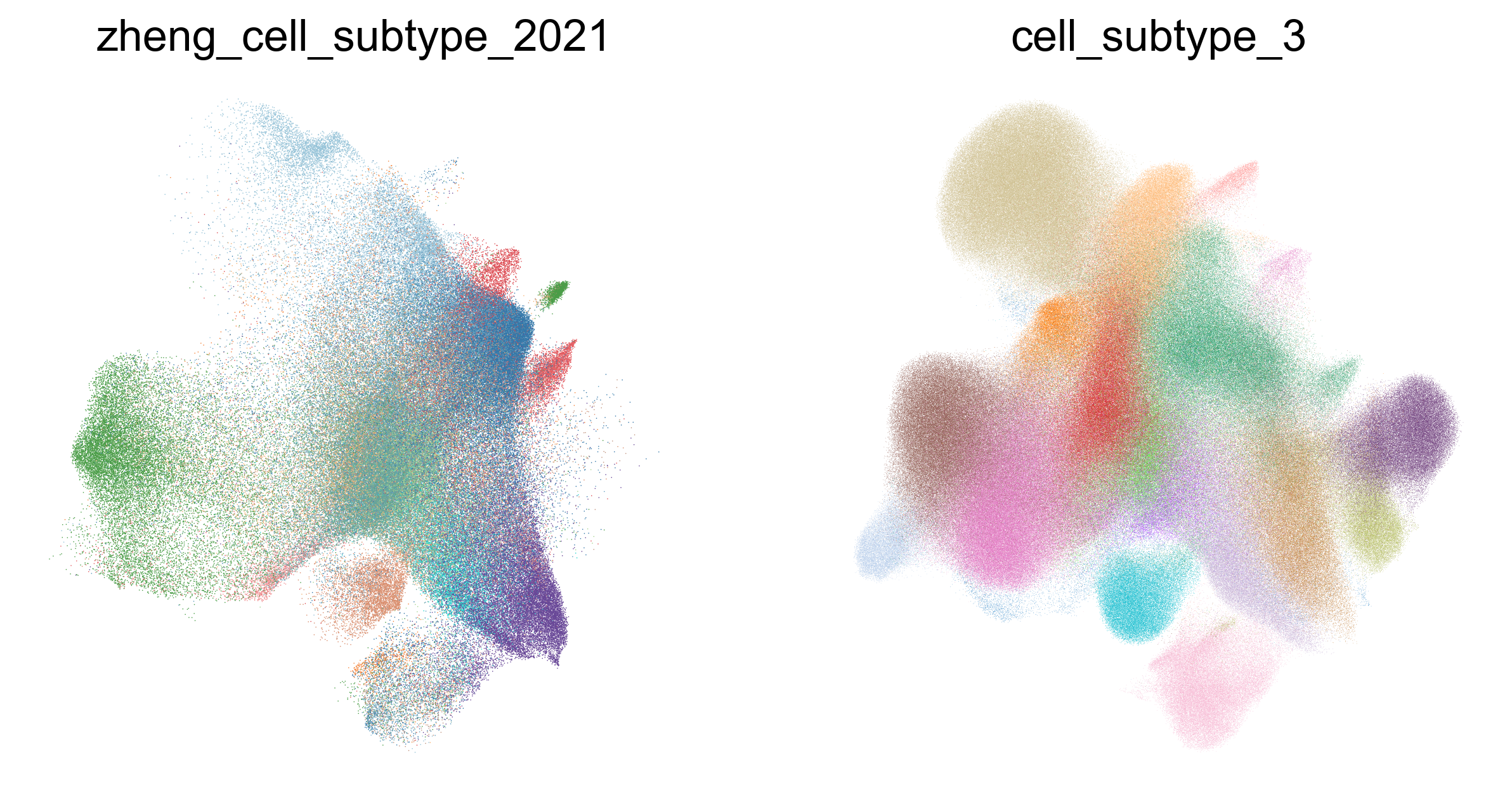
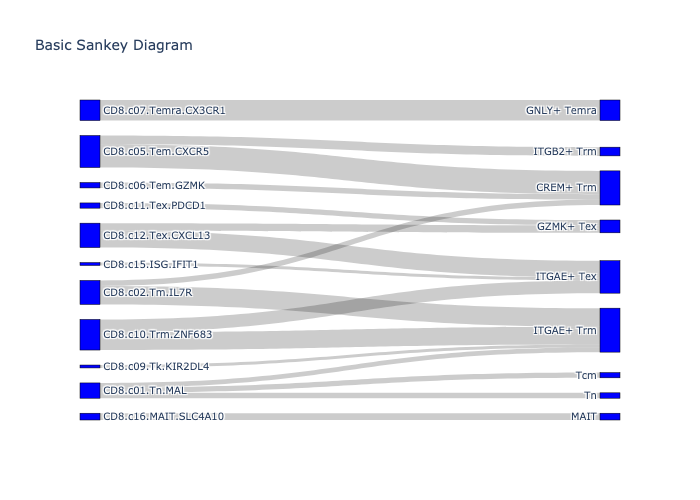
_, fig = scatlasvae.tl.cell_type_alignment(
adata_cd8_zheng_2021_merged,
obs_1 = 'cell_subtype_zheng_2021',
obs_2 = 'cell_subtype_3_prediction',
perc_in_obs_1 = 0.2
)
Cross-atlas integration between pan-disease CD8 atlas and Chu et al., 2023 TCellMap¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
adata_cd8_chu_2023.obs['cell_subtype_3'] = 'undefined'
adata_cd8.obs['cell_subtype_3'] = list(pd.read_csv("/rsch/Snowxue/2023-NM-Revision-GEX-HuARdb-Notebooks/huARdb_v2_GEX.CD8.hvg4k.20231116.obs.cell_subtype_3.csv")['cell_subtype_3'])
adata_cd8.obs['label_4'] = 'undefined'
adata_cd8.obs['atlas_name'] = 'huARdbv2'
adata_cd8_chu_2023_merged = sc.concat([
adata_cd8_chu_2023, adata_cd8
], join='inner')
adata_cd8_chu_2023_merged.obs['cell_subtype_3'] = list(adata_cd8_chu_2023_merged.obs['cell_subtype_3'])
adata_cd8_chu_2023_merged.obs['cell_subtype_3'] = adata_cd8_chu_2023_merged.obs['cell_subtype_3'].fillna("undefined")
multi_atlas_vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8_chu_2023_merged,
batch_key='sample_name',
additional_batch_keys=['study_name','atlas_name'],
batch_embedding='embedding',
batch_hidden_dim=64,
label_key='cell_subtype_3',
additional_label_keys=['cell_subtype_chu_2023'],
device='cuda:2'
)
_ = multi_atlas_vae_model.fit(
max_epoch=32,
lr=5e-5,
kl_weight=1.,
n_epochs_kl_warmup=16
)
import umap
adata_cd8_chu_2023_merged.obsm['X_gex'] = multi_atlas_vae_model.get_latent_embedding()
adata_cd8_chu_2023_merged.obsm['X_umap'] = umap.UMAP().fit_transform(adata_cd8_chu_2023_merged.obsm['X_gex'])
predictions = multi_atlas_vae_model.predict_labels(return_pandas=True)
predictions_logits = multi_atlas_vae_model.predict_labels(return_pandas=False)
adata_cd8_chu_2023_merged.uns['cell_subtype_3_prediction_logits'] = predictions_logits[0].detach().cpu().numpy()
adata_cd8_chu_2023_merged.uns['cell_subtype_chu_2023_prediction_logits'] = predictions_logits[1][0].detach().cpu().numpy()
adata_cd8_chu_2023_merged.obs['cell_subtype_3_prediction'] = list(predictions['cell_subtype_3'])
adata_cd8_chu_2023_merged.obs['cell_subtype_chu_2023_prediction'] = list(predictions['cell_subtype_chu_2023'])
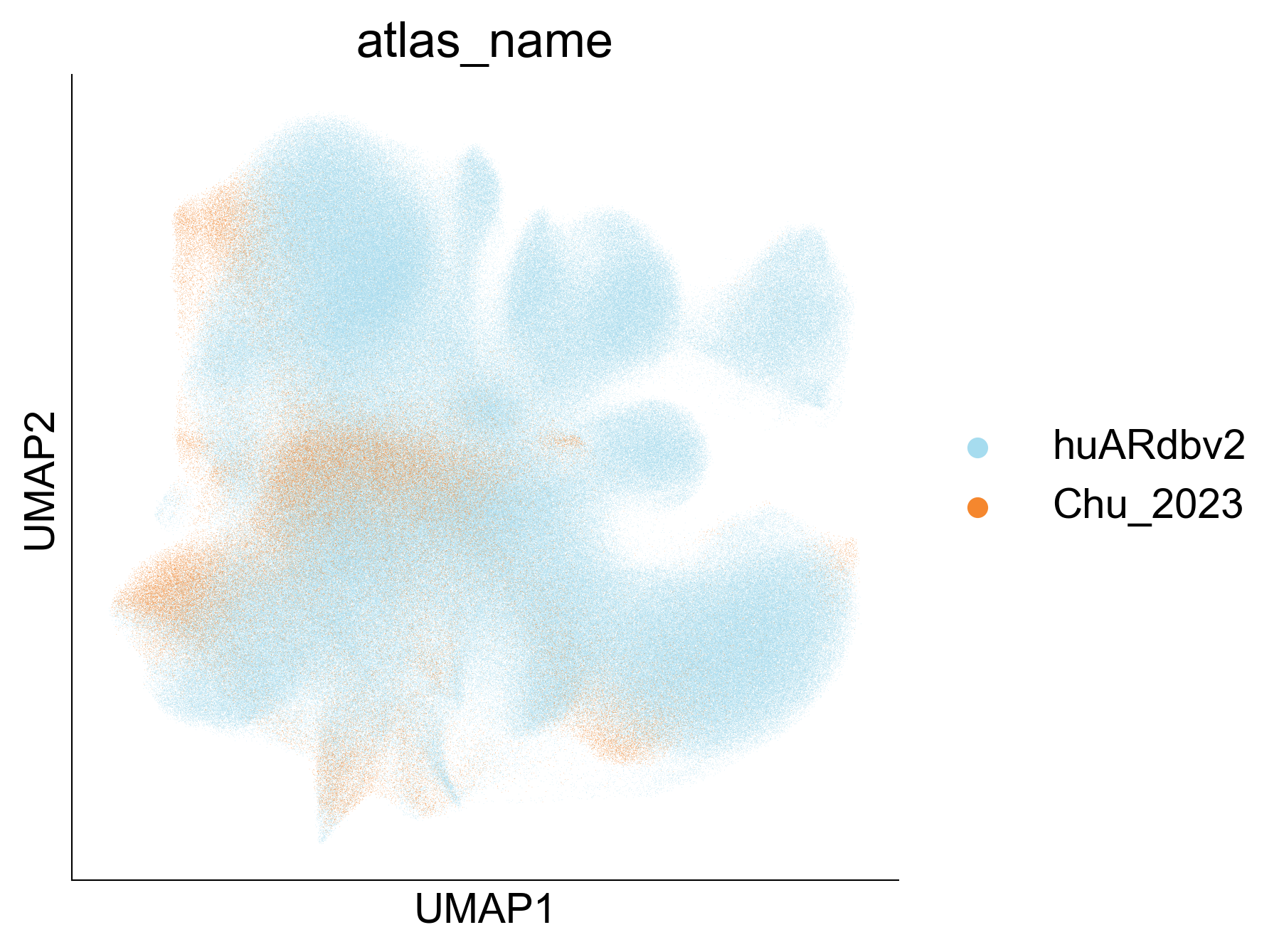
Extended Data Figure 9B¶
fig,ax=createFig(figsize=(5,5))
sc.pl.umap(adata_cd8_chu_2023_merged[adata_cd8_chu_2023_merged.obs['atlas_name'] == 'huARdbv2'], color='atlas_name', ax=ax, palette=
{
'Chu_2023': '#F5872E',
'huARdbv2': '#A6DCEF'
}, show=False, s=0.1
)
sc.pl.umap(adata_cd8_chu_2023_merged[adata_cd8_chu_2023_merged.obs['atlas_name'] != 'huARdbv2'], color='atlas_name', ax=ax, palette=
{
'Chu_2023': '#F5872E',
'huARdbv2': '#A6DCEF'
}, show=False, s=0.1
)
fig.savefig("/Users/snow/Desktop/tmp.png")
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning:
Trying to modify attribute `._uns` of view, initializing view as actual.
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning:
X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning:
No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning:
Trying to modify attribute `._uns` of view, initializing view as actual.
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning:
X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning:
No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
fig,ax=createSubplots(1,2,figsize=(10,5))
obsm = adata_cd8_chu_2023_merged.obsm['X_umap']
sc.pl.umap(adata_cd8_chu_2023_merged[
adata_cd8_chu_2023_merged.obs['cell_subtype_chu_2023'] != 'undefined'
], color='cell_subtype_chu_2023', ax=ax[0], show=False, palette=chu_annotation_cmap_2, frameon=False, legend_loc='none')
sc.pl.umap(adata_cd8_chu_2023_merged[
adata_cd8_chu_2023_merged.obs['cell_subtype_3'] != 'undefined'
], color='cell_subtype_3', ax=ax[1], palette=subtype_color, frameon=False, legend_loc='none')
fig.savefig("/Users/snow/Desktop/tmp.png")
plt.show()
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
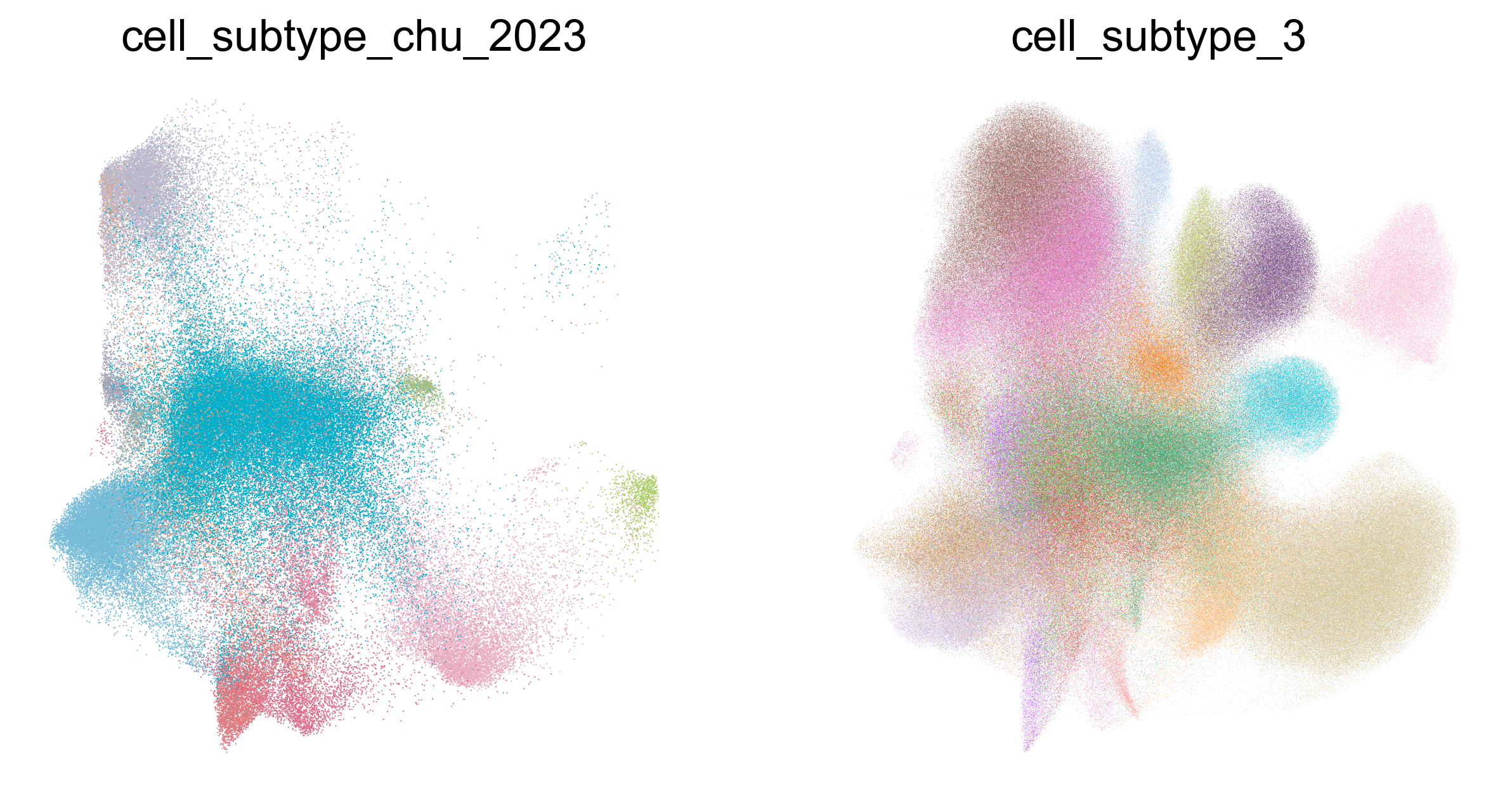
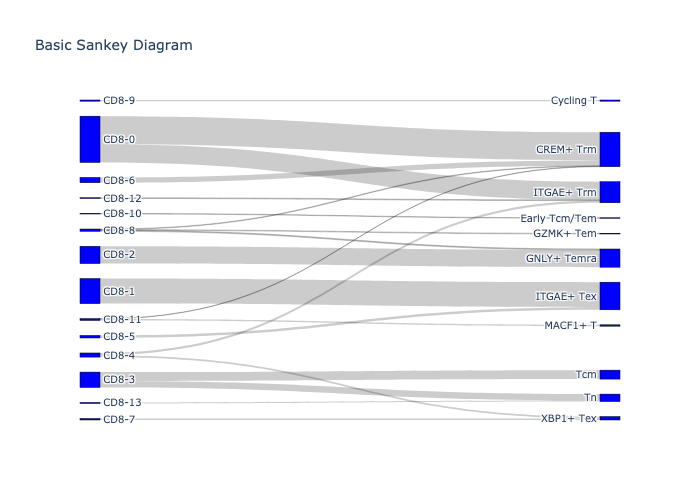
_, fig = scatlasvae.tl.cell_type_alignment(
adata_cd8_chu_2023_merged,
obs_1 = 'cell_subtype_chu_2023',
obs_2 = 'cell_subtype_3_prediction',
perc_in_obs_1 = 0.2
)
Cross-atlas integration of Tex subset between pan-disease CD8 atlas and pan-cancer CD8 T landscape¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
# Load data from Zheng et al., 2021
adata_cd8_zheng_2021 = sc.read_h5ad("../data/adata_cd8_zheng_2021.h5ad")
adata_cd8_zheng_2021_tex_tpex = adata_cd8_zheng_2021[
adata_cd8_zheng_2021.obs["cell_subtype_zheng_2021"].isin(
[
"CD8.c11.Tex.PDCD1",
"CD8.c12.Tex.CXCL13",
"CD8.c14.Tex.TCF7",
"CD8.c15.ISG.IFIT1",
]
)
]
adata_cd8_tex = adata_cd8[
adata_cd8.obs['cell_subtype_3'].isin([
'GZMK+ Tex',
'ITGAE+ Tex',
'XBP1+ Tex'
])
]
adata_cd8_zheng_2021_tex_tpex.obs["cell_subtype_3"] = "undefined"
adata_cd8_tex.obs["cell_subtype_zheng_2021"] = "undefined"
adata_cd8_tex.obs["atlas_name"] = "huARdbv2"
adata_cd8_tex_tpex_merged = sc.concat(
[adata_cd8_zheng_2021_tex_tpex, adata_cd8_tex], join="inner"
)
adata_cd8_tex_tpex_merged.obs["cell_subtype_3"] = list(
adata_cd8_tex_tpex_merged.obs["cell_subtype_3"]
)
adata_cd8_tex_tpex_merged.obs["cell_subtype_3"] = adata_cd8_tex_tpex_merged.obs[
"cell_subtype_3"
].fillna("undefined")
# Using GPU #2
multi_atlas_vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8_tex_tpex_merged,
batch_key="sample_name",
additional_batch_keys=["study_name", "atlas_name"],
batch_embedding="embedding",
batch_hidden_dim=64,
label_key="cell_subtype_3",
additional_label_keys=["cell_subtype_zheng_2021"],
device="cuda:2",
)
multi_atlas_vae_model.fit(
max_epoch=64,
lr=5e-5,
kl_weight=1.,
n_epochs_kl_warmup=32
)
Figure 3F¶
fig,ax=createFig(figsize=(5,5))
sc.pl.umap(adata_cd8_tex_tpex_merged, color='atlas_name', ax=ax, palette=
{
'huARdbv2': '#A6DCEF',
'zheng_2021': '#284DA1'
}, show=False, s=1
)
plt.show()
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
zheng_2021_annotation_cmap_cd8 = {
"CD8.c01.Tn.MAL": "#96C3D8",
"CD8.c02.Tm.IL7R": "#5D9BBE",
"CD8.c03.Tm.RPS12": "#F5B375",
"CD8.c04.Tm.CD52": "#C0937E",
"CD8.c05.Tem.CXCR5": "#67A59B",
"CD8.c06.Tem.GZMK": "#A4D38E",
"CD8.c07.Temra.CX3CR1": "#4A9D47",
"CD8.c08.Tk.TYROBP": "#F19294",
"CD8.c09.Tk.KIR2DL4": "#E45A5F",
"CD8.c10.Trm.ZNF683": "#3477A9",
"CD8.c11.Tex.PDCD1": "#BDA7CB",
"CD8.c12.Tex.CXCL13": "#684797",
"CD8.c13.Tex.myl12a": "#9983B7",
"CD8.c14.Tex.TCF7": "#CD9A99",
"CD8.c15.ISG.IFIT1": "#DD4B52",
"CD8.c16.MAIT.SLC4A10": "#DA8F6F",
"CD8.c17.Tm.NME1": "#F58135",
}
zheng_2021_annotation_cmap_cd8["CD8.c11.Tex.PDCD1"] = "#21D5CE"
fig, ax = createSubplots(1, 2, figsize=(10, 5))
sc.pl.umap(
adata_cd8_tex_tpex_merged[
adata_cd8_tex_tpex_merged.obs["cell_subtype_zheng_2021"] != "undefined"
],
color="cell_subtype_zheng_2021",
ax=ax[1],
show=False,
palette=zheng_2021_annotation_cmap_cd8,
frameon=False,
legend_loc="none",
)
sc.pl.umap(
adata_cd8_tex_tpex_merged[
adata_cd8_tex_tpex_merged.obs["cell_subtype"] != "undefined"
],
color="cell_subtype",
ax=ax[0],
palette=subtype_color,
frameon=False,
show=False,
legend_loc="none",
)
plt.show()
fig.savefig("/Users/snow/Desktop/tmp.png")
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(

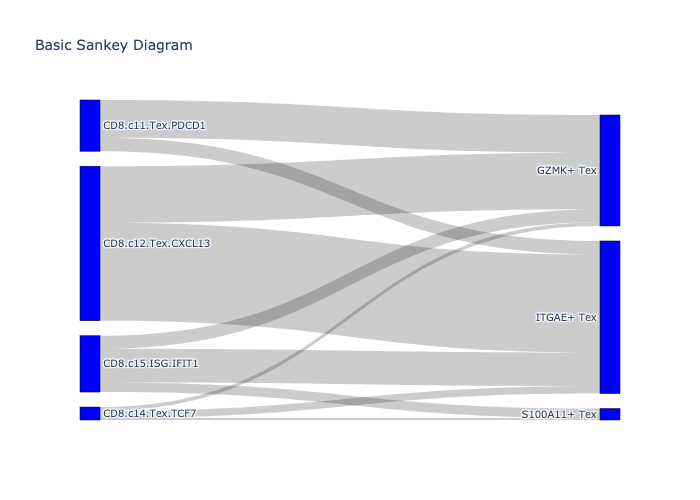
Figure 3G¶
We use cell type alignment from the scatlasvae package
_, fig = scatlasvae.tl.cell_type_alignment(
adata_cd8_tex_tpex_merged,
obs_1='cell_subtype_zheng_2021',
obs_2='cell_subtype_3_prediction'
)
fig.show(renderer="png")
Supervised integration of pan-disease CD8 T atlas¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8,
batch_key="sample_name",
additional_batch_keys=["study_name"],
batch_embedding="embedding",
batch_hidden_dim=64,
constrain_latent_embedding=True,
label_key="cell_subtype_3",
)
vae_model.fit(max_epoch=10, lr=5e-5)
vae_model.save_to_disk("./huARdb_v2_GEX.CD8.hvg4k.supervised.model")
Retrainined MAIT/ILTCK subset for clustering¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
adata_cd8_iltck_mait = adata_cd8[
adata_cd8.obs['cell_subtype_3'].isin([
'MAIT','ILTCK'
])
]
import umap
vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8_iltck_mait,
batch_key="sample_name",
additional_batch_keys=["study_name"],
batch_embedding="embedding",
batch_hidden_dim=10,
device="cuda:0",
)
result = vae_model.fit(max_epoch=32, n_epochs_kl_warmup=16, lr=5e-5, kl_weight=3.0)
adata_cd8_iltck_mait.obsm["X_retrain_gex"] = vae_model.get_latent_embedding()
adata_cd8_iltck_mait.obsm["X_retrain_umap"] = umap.UMAP().fit_transform(
adata_cd8_iltck_mait.obsm["X_retrain_gex"]
)
import scanpy as sc
adata_mait_iltck = sc.read_h5ad("/Users/snow/Desktop/2023-NM-Revision-GEX-HuARdb-Notebooks/data/huARdb_v2_GEX.CD8.hvg4k.20231116.ILTCK_MAIT.vae.h5ad")
tcr_df = pd.read_csv("/Users/snow/Desktop/2023-NM-Revision-GEX-HuARdb-Notebooks/data/huARdb_v2_GEX.CD8.hvg4k.TCR.csv", index_col=0)
adata_mait_iltck.obs = adata_mait_iltck.obs.join(tcr_df.loc[adata_mait_iltck.obs.index])
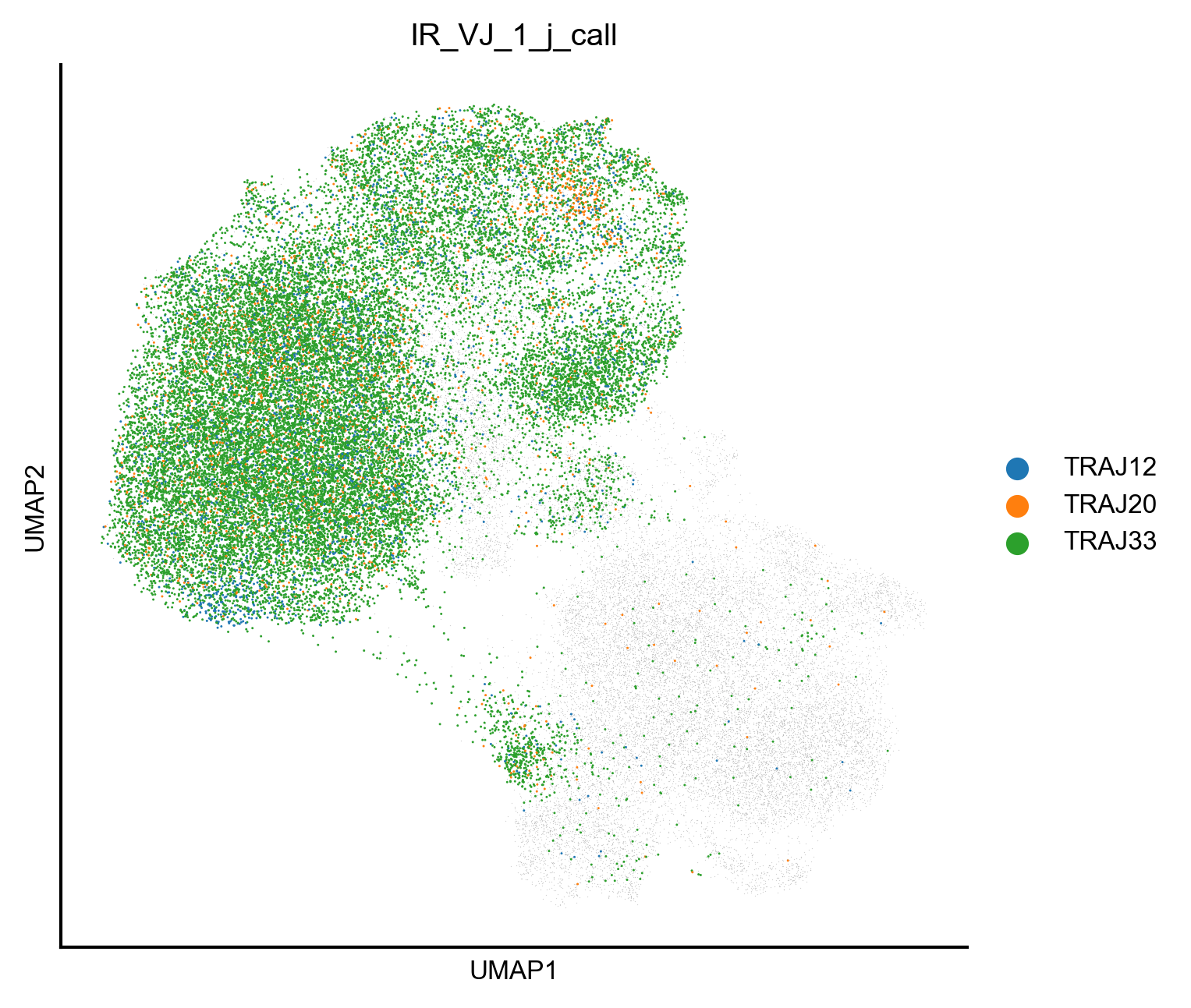
Extended Data Figure 8i¶
fig,ax=createFig(figsize=(5,5))
import scanpy as sc
obsm = adata_mait_iltck.obsm['X_umap']
ax.scatter(
obsm[:,0],
obsm[:,1],
lw=0,
c='#A7A7A7',
s=0.1,
alpha=0.5
)
sc.pl.umap(
adata_mait_iltck[
np.array(adata_mait_iltck.obs['IR_VJ_1_v_call'] == 'TRAV1-2') & (
np.array(adata_mait_iltck.obs['IR_VJ_1_j_call'] == 'TRAJ33') |
np.array(adata_mait_iltck.obs['IR_VJ_1_j_call'] == 'TRAJ20') |
np.array(adata_mait_iltck.obs['IR_VJ_1_j_call'] == 'TRAJ12')
)
],
color='IR_VJ_1_j_call',
ax=ax,
s=2,
layer='normalized'
)
/usr/local/lib/python3.9/site-packages/anndata/_core/anndata.py:1235: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
df[key] = c
/usr/local/Cellar/python@3.9/3.9.18_1/Frameworks/Python.framework/Versions/3.9/lib/python3.9/contextlib.py:126: FutureWarning: X.dtype being converted to np.float32 from int32. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
next(self.gen)
/usr/local/lib/python3.9/site-packages/anndata/_core/anndata.py:1235: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
df[key] = c
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
fig,ax=createFig(figsize=(5,5))
import scanpy as sc
obsm = adata_mait_iltck.obsm['X_umap']
sc.pl.umap(
adata_mait_iltck,
color='study_name',
ax=ax,
s=2,
layer='normalized',
cmap='Reds'
)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
Extended Data Figure 8h¶
fig,axes=createSubplots(2,2,figsize=(8,8))
axes=axes.flatten()
import scanpy as sc
obsm = adata_mait_iltck.obsm['X_umap']
sc.pl.umap(
adata_mait_iltck,
color='cell_subtype_3',
ax=axes[0],
s=1,
show=False
)
sc.pl.umap(
adata_mait_iltck,
color='TYROBP',
ax=axes[1],
s=1,
show=False,
layer='normalized',
cmap='Reds'
)
sc.pl.umap(
adata_mait_iltck,
color='S100A11',
ax=axes[2],
s=1,
show=False,
layer='normalized',
cmap='Reds'
)
sc.pl.umap(
adata_mait_iltck,
color='FCER1G',
ax=axes[3],
s=1,
show=True,
layer='normalized',
cmap='Reds'
)
plt.show()
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
Retraining Tex subset for higher resolution clustering¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
adata_cd8_tex = adata_cd8[
adata_cd8.obs['cell_subtype_3'].isin([
'GZMK+ Tex',
'ITGAE+ Tex',
'XBP1+ Tex'
])
]
import umap
vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8_tex,
batch_key="sample_name",
additional_batch_keys=["study_name"],
batch_embedding="embedding",
batch_hidden_dim=10,
device="cuda:0",
)
result = vae_model.fit(max_epoch=32, n_epochs_kl_warmup=16, lr=5e-5, kl_weight=3.0)
adata_cd8_tex.obsm["X_retrain_gex"] = vae_model.get_latent_embedding()
adata_cd8_tex.obsm["X_retrain_umap"] = umap.UMAP().fit_transform(
adata_cd8_tex.obsm["X_retrain_gex"]
)
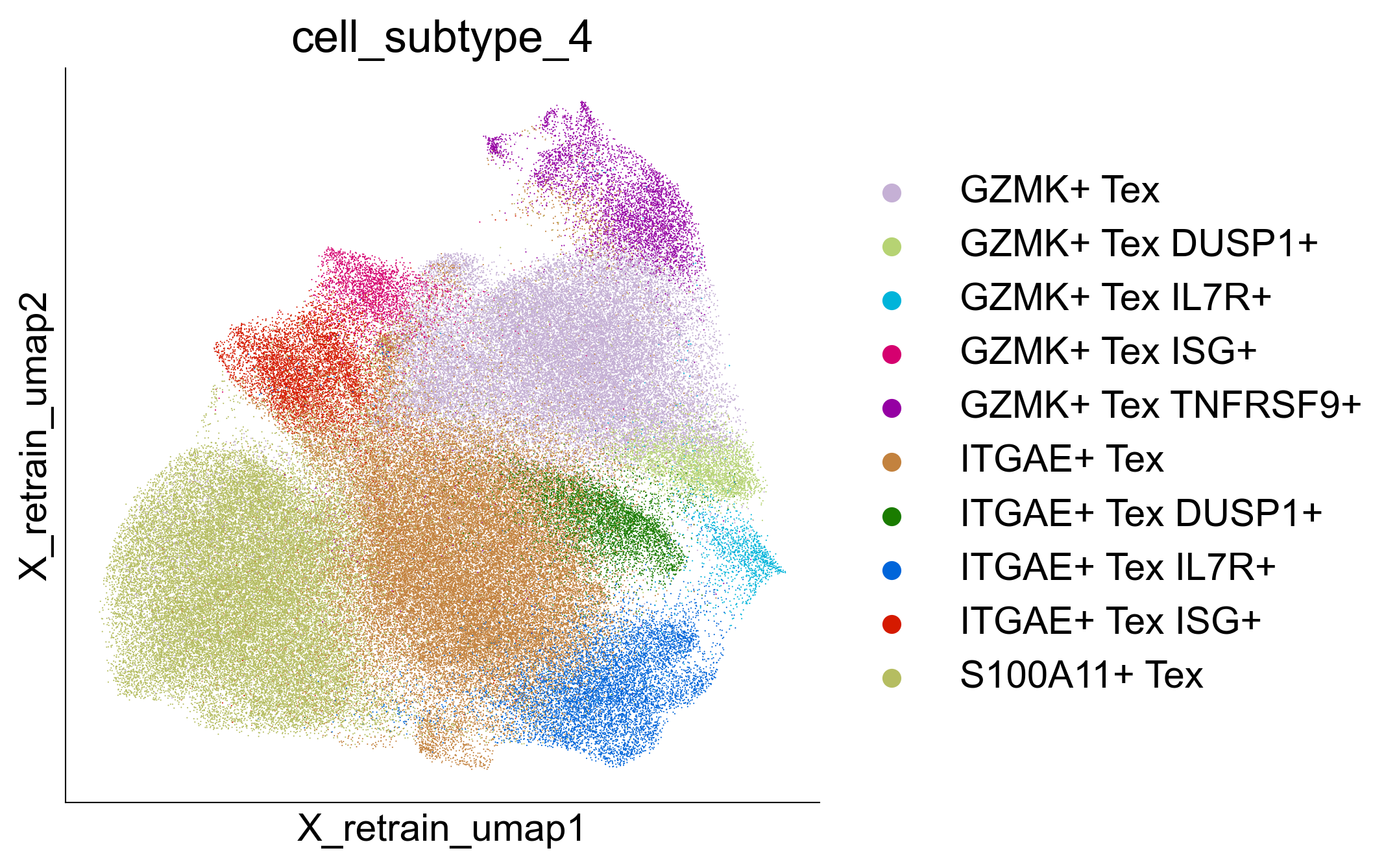
Extended Data Figure 10G¶
fig, ax = createFig(figsize=(5, 5))
sc.pl.embedding(
adata_cd8_tex,
color="cell_subtype_4",
ax=ax,
basis="X_retrain_umap",
layer="normalized",
cmap="Reds",
s=1,
)
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning:
No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
<Figure size 1800x1200 with 0 Axes>
Supervised integration of the Tex subset¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
tex_vae_model = scatlasvae.model.scAtlasVAE(
adata=adata_cd8_tex,
batch_key="sample_name",
additional_batch_keys=["study_name"],
batch_embedding="embedding",
batch_hidden_dim=10,
label_key="cell_subtype_4",
device="cuda:2",
constrain_latent_key="X_gex_retrained_on_4k_hvg",
constrain_latent_embedding=True,
)
tex_vae_model.fit(
max_epoch=32, lr=5e-5, kl_weight=1.0, n_epochs_kl_warmup=10, pred_last_n_epoch=22
)
tex_vae_model.save_to_disk("./huARdb_v2_GEX.CD8.hvg4k.Tex.supervised.model")
Query-to-reference mapping to pan-disease CD8 T atlas¶
Loading query data¶
adata_bassez_cohort1 = sc.read_h5ad("./transfer_data/Bassez_BC.cohort1.CD8_T.h5ad")
adata_bassez_cohort2 = sc.read_h5ad("./transfer_data/Bassez_BC.cohort2.CD8_T.h5ad")
adata_bi = sc.read_h5ad("./transfer_data/Bi_RCC.CD8_T.h5ad")
adata_caushi = sc.read_h5ad('./transfer_data/Caushi_NSCLC.CD8_T.h5ad')
adata_liu = sc.read_h5ad("./transfer_data/Liu_TNBC.CD8_T.h5ad")
adata_luoma_pbmc = sc.read_h5ad("./transfer_data/Luoma_HNSCC_PBMC.CD8_T.h5ad")
adata_luoma_til = sc.read_h5ad("./transfer_data/Luoma_HNSCC_TIL.CD8_T.h5ad")
adata_watson = sc.read_h5ad("./transfer_data/Watson_MELA.CD8_T.h5ad")
adata_zhang = sc.read_h5ad("./transfer_data/Zhang_LC.CD8_T.h5ad")
merged_adata_for_transfer = sc.concat([adata_bassez_cohort1,
adata_bassez_cohort2,
adata_bi,
adata_caushi,
adata_liu,
adata_luoma_pbmc,
adata_luoma_til,
adata_watson,
adata_zhang
])
Transfer using pretrained models¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
## ALL SCRIPT
adata_transfer = sc.read_h5ad("../Merged_adata_for_transfer.3.h5ad")
state_dict = torch.load("./huARdb_v2_GEX.CD8.hvg4k.supervised.model")
adata_transfer.obs["cell_subtype_3"] = "undefined"
adata_transfer = adata_transfer[:, adata_cd8.var.index]
adata_transfer.obs["cell_subtype_3"] = pd.Categorical(
list(adata_transfer.obs["cell_subtype_3"]),
categories=pd.Categorical(adata_cd8.obs["cell_subtype_3"]).categories,
)
adata_transfer.obs["study_name"] = "Lechner_2023"
vae_model_transfer = scatlasvae.model.scAtlasVAE(
adata=adata_transfer,
pretrained_state_dict=state_dict["model_state_dict"],
**state_dict["model_config"]
)
adata_transfer.obsm["X_gex"] = vae_model_transfer.get_latent_embedding()
adata_transfer.obsm["X_umap"] = scatlasvae.tl.transfer_umap(
adata_cd8.obsm["X_gex"],
adata_cd8.obsm["X_umap"],
adata_transfer.obsm["X_gex"],
method="knn",
n_neighbors=3,
)["embedding"]
df = vae_model_transfer.predict_labels(return_pandas=True)
adata_transfer.obs["cell_subtype_3"] = list(df["cell_subtype_3"])
import torch
adata_transfer_tex = adata_transfer[
list(map(lambda x: "Tex" in x, adata_transfer.obs["cell_subtype_3"]))
]
adata_transfer_tex.obs["cell_subtype_4"] = "undefined"
adata_transfer_tex.obs["cell_subtype_4"] = pd.Categorical(
list(adata_transfer_tex.obs["cell_subtype_4"]),
categories=adata_cd8_tex.obs["cell_subtype_4"].cat.categories,
)
state_dict = torch.load(
"huARdb_v2_GEX.CD8.hvg4k.Tex.supervised.model"
)
vae_model_transfer = scatlasvae.model.scAtlasVAE(
adata=adata_transfer_tex,
pretrained_state_dict=state_dict["model_state_dict"],
**state_dict["model_config"]
)
adata_transfer_tex.obsm["X_retrain_GEX"] = vae_model_transfer.get_latent_embedding()
adata_transfer_tex.obsm["X_retrain_umap"] = scatlasvae.tl.transfer_umap(
adata_cd8_tex.obsm["X_retrain_GEX"],
adata_cd8_tex.obsm["X_retrain_umap"],
adata_transfer_tex.obsm["X_retrain_GEX"],
method="knn",
n_neighbors=3,
)["embedding"]
adata_transfer_tex.obs["cell_subtype_4"] = vae_model_transfer.predict_labels(
return_pandas=True
)["cell_subtype_4"]
fig,ax=createFig(figsize=(5,5))
obsm = adata_cd8.obsm['X_umap']
obsm = obsm[np.random.choice(list(range(len(adata_cd8))), 100000, replace=False)]
ax.scatter(
obsm[:,0],
obsm[:,1],
lw=0,
c='#A7A7A7',
s=0.1,
alpha=0.5
)
sc.pl.umap(adata_transfer, color='cell_subtype_3', ax=ax, palette=subtype_color, layer='normalized')
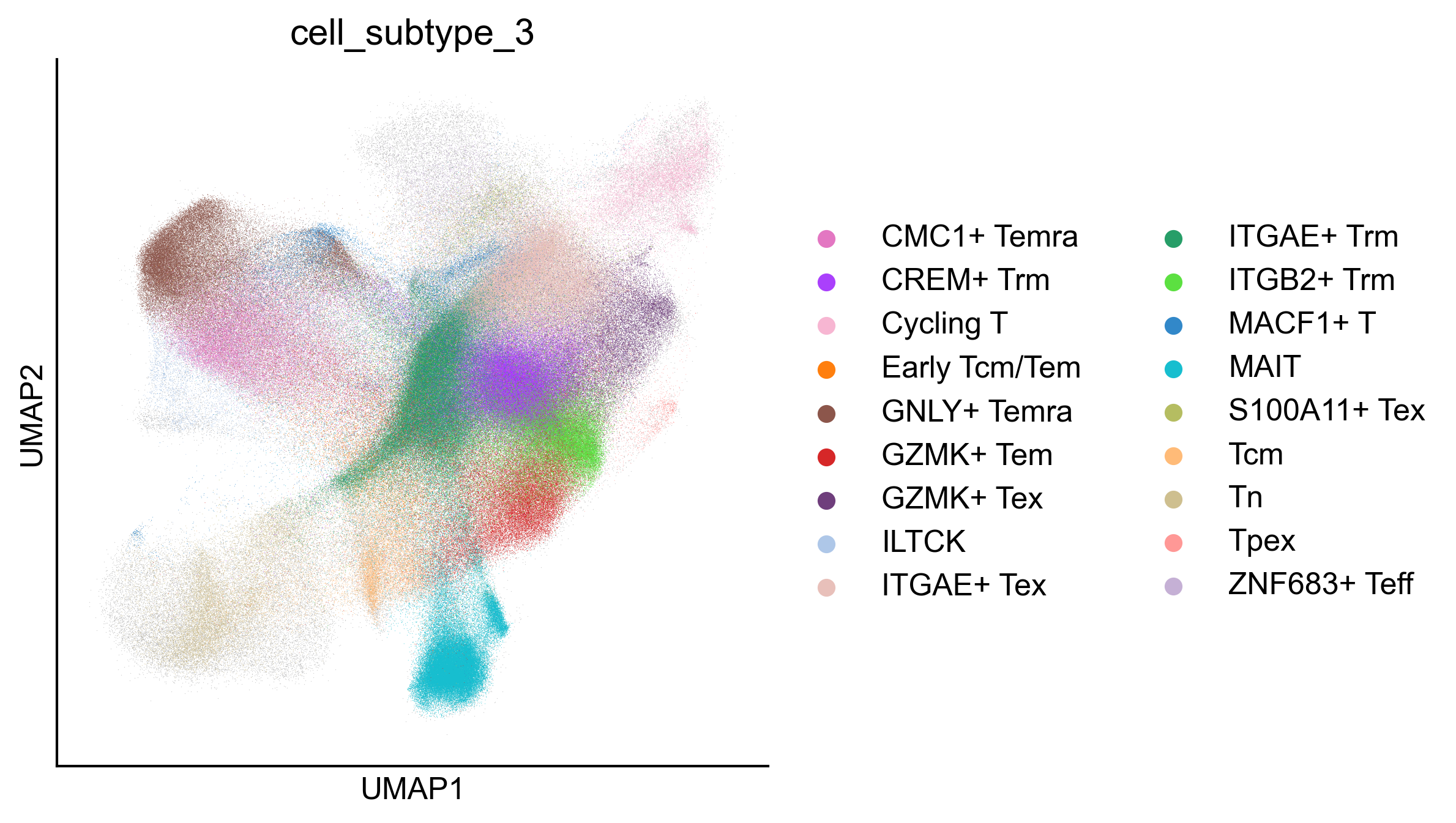
Figure 5A¶
fig, ax = createFig(figsize=(5, 5))
ax.scatter(obsm[:, 0], obsm[:, 1], lw=0, s=0.2, color="#F7F7F7")
sc.pl.umap(
merged_adata_for_transfer[
merged_adata_for_transfer.obs["study_name"] != "Smillie_2019"
],
color="study_name",
# palette=subtype_color,
palette=sc.pl.palettes.godsnot_102[56:61] + sc.pl.palettes.godsnot_102[62:],
ax=ax,
s=0.5,
alpha=0.6,
)
fig.savefig("./figures/merged_adata_for_transfer.zero_shot_transfer.cell_subtype_3.png")
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/anndata/_core/anndata.py:1828: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
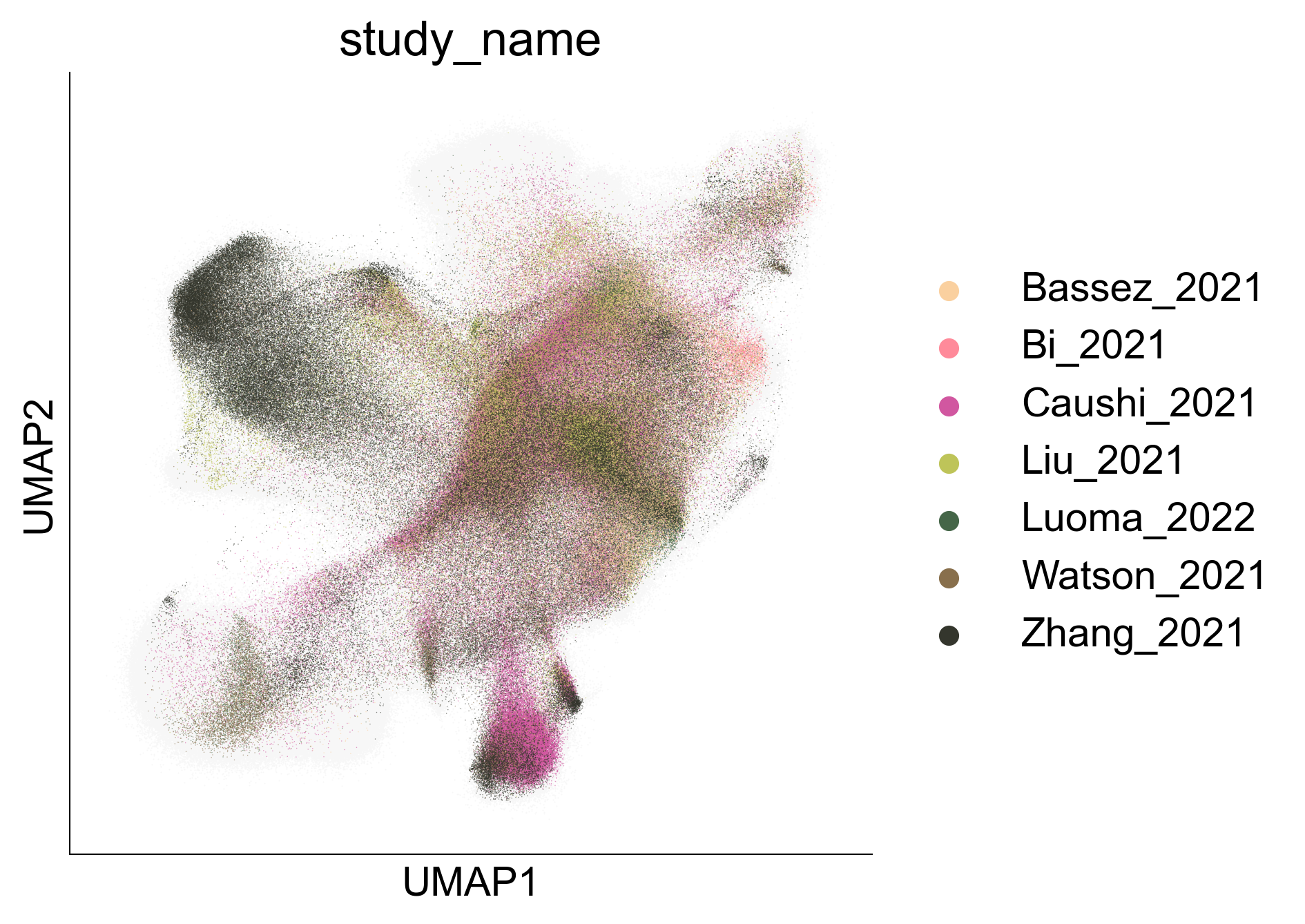
fig, ax = createFig(figsize=(5, 5))
sc.pl.embedding(
adata_transfer_tex, color="cell_subtype_4", ax=ax, basis="X_retrain_umap"
)
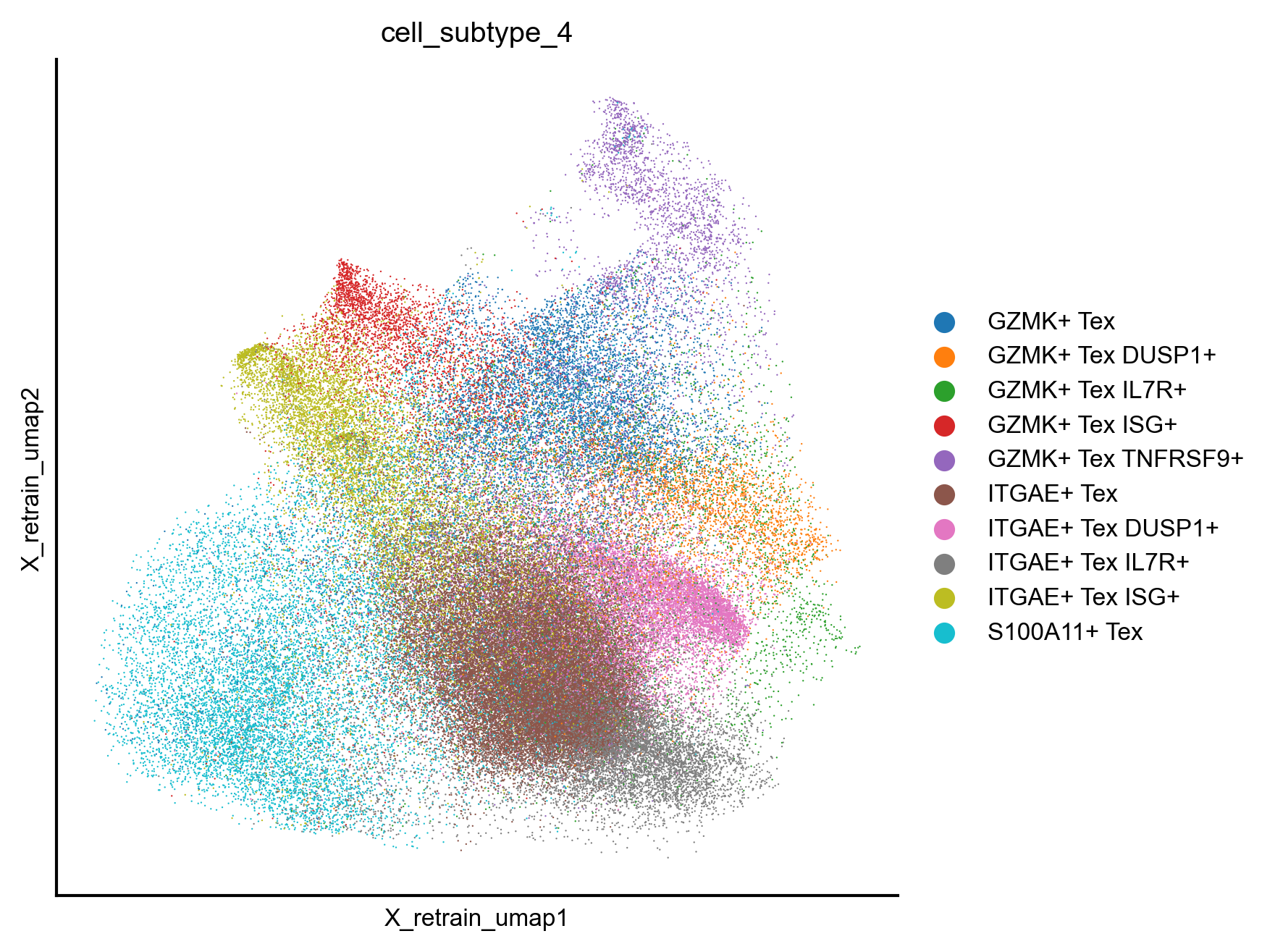
Figure 5B¶
sc.pl.dotplot(
merged_adata_for_transfer,
var_names=[
"S1PR1",
"LEF1",
"SELL",
"IL7R",
"KLRG1",
"GZMK",
"GZMB",
"GNLY",
"ZNF683",
"KLRB1",
"ZBTB16",
"FCER1G",
"TYROBP",
"CMC1",
"ITGAE",
"CREM",
"TCF7",
"PDCD1",
"LAG3",
"HAVCR2",
"CXCR6",
"TIGIT",
"XBP1",
"MACF1",
"MKI67",
"CDC20",
],
groupby="cell_subtype_3",
dot_max=0.7,
standard_scale="var",
layer="normalized",
save="20231220_Merged_adata_for_transfer.zero_shot_transfer",
)
WARNING: saving figure to file figures/dotplot_20231220_Merged_adata_for_transfer.zero_shot_transfer.pdf
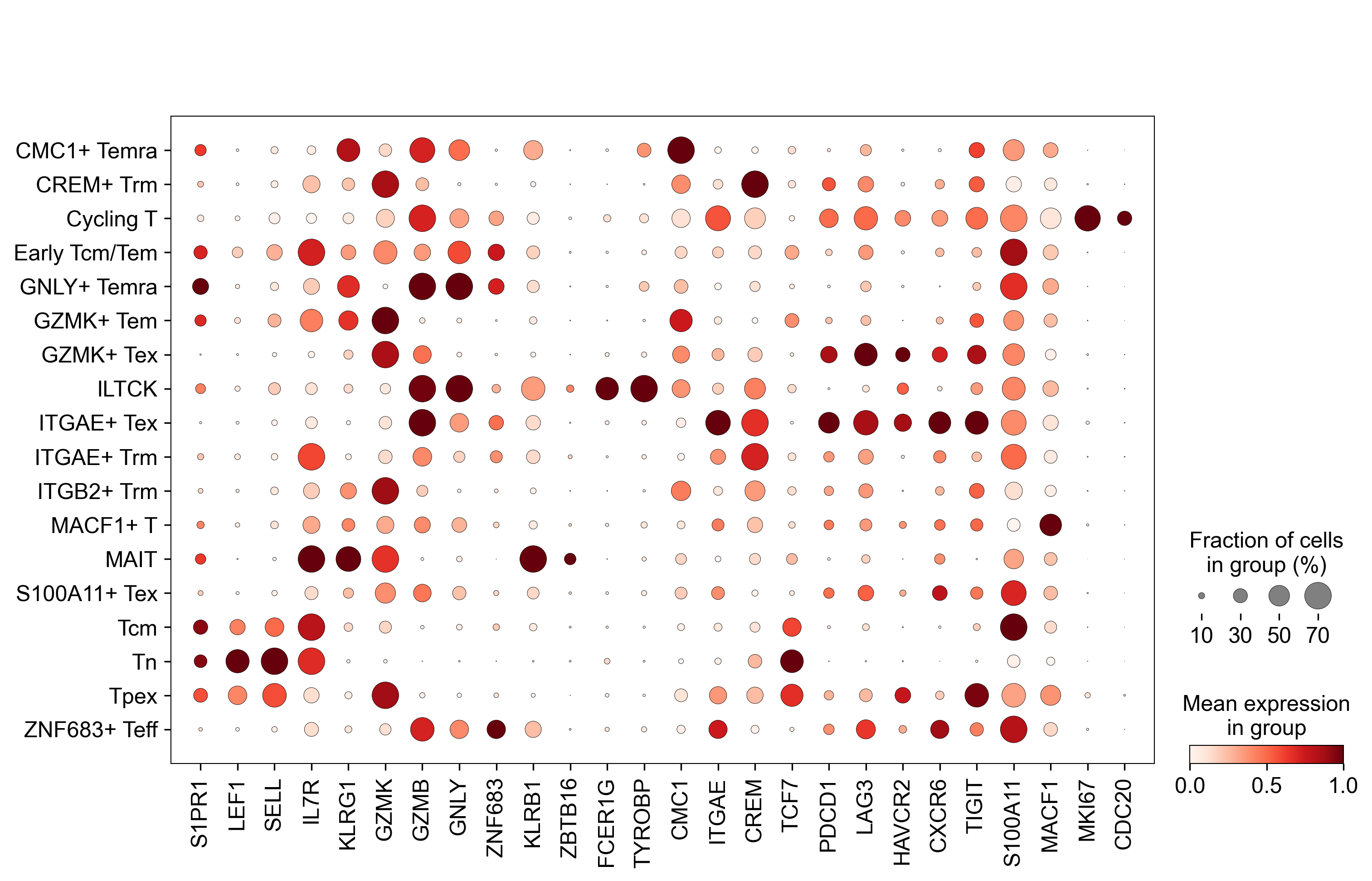
Figure 5C¶
fig, axes = createSubplots(3, 3, figsize=(10, 10))
axes = axes.flatten()
for ax in axes:
ax.scatter(obsm[:, 0], obsm[:, 1], lw=0, s=0.2, color="#F7F7F7")
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Watson_2021")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "PBMC")
],
color="cell_subtype_3",
palette=subtype_color,
show=False,
ax=axes[0],
s=3,
legend_loc="none",
)
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Luoma_2022")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "PBMC")
],
color="cell_subtype_3",
palette=subtype_color,
ax=axes[1],
s=3,
alpha=0.6,
show=False,
legend_loc="none",
)
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Zhang_2021")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "PBMC")
],
color="cell_subtype_3",
palette=subtype_color,
ax=axes[2],
s=3,
alpha=0.6,
show=False,
legend_loc="none",
)
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Bassez_2021")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "TIL")
],
color="cell_subtype_3",
palette=subtype_color,
show=False,
ax=axes[3],
s=3,
alpha=0.6,
legend_loc="none",
)
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Luoma_2022")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "TIL")
],
color="cell_subtype_3",
palette=subtype_color,
ax=axes[4],
s=3,
alpha=0.6,
show=False,
legend_loc="none",
)
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Liu_2021")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "TIL")
],
color="cell_subtype_3",
palette=subtype_color,
ax=axes[5],
s=3,
alpha=0.6,
show=False,
legend_loc="none",
)
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Zhang_2021")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "TIL")
],
color="cell_subtype_3",
palette=subtype_color,
ax=axes[6],
s=3,
alpha=0.6,
show=False,
legend_loc="none",
)
sc.pl.umap(
merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["study_name"] == "Caushi_2021")
& np.array(merged_adata_for_transfer.obs["tissue_type"] == "TIL")
],
color="cell_subtype_3",
palette=subtype_color,
ax=axes[7],
s=3,
alpha=0.6,
show=False,
legend_loc="none",
)
plt.show()
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
/usr/local/lib/python3.9/site-packages/anndata/compat/_overloaded_dict.py:106: ImplicitModificationWarning: Trying to modify attribute `._uns` of view, initializing view as actual.
self.data[key] = value
/usr/local/lib/python3.9/site-packages/anndata/_core/anndata.py:1828: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
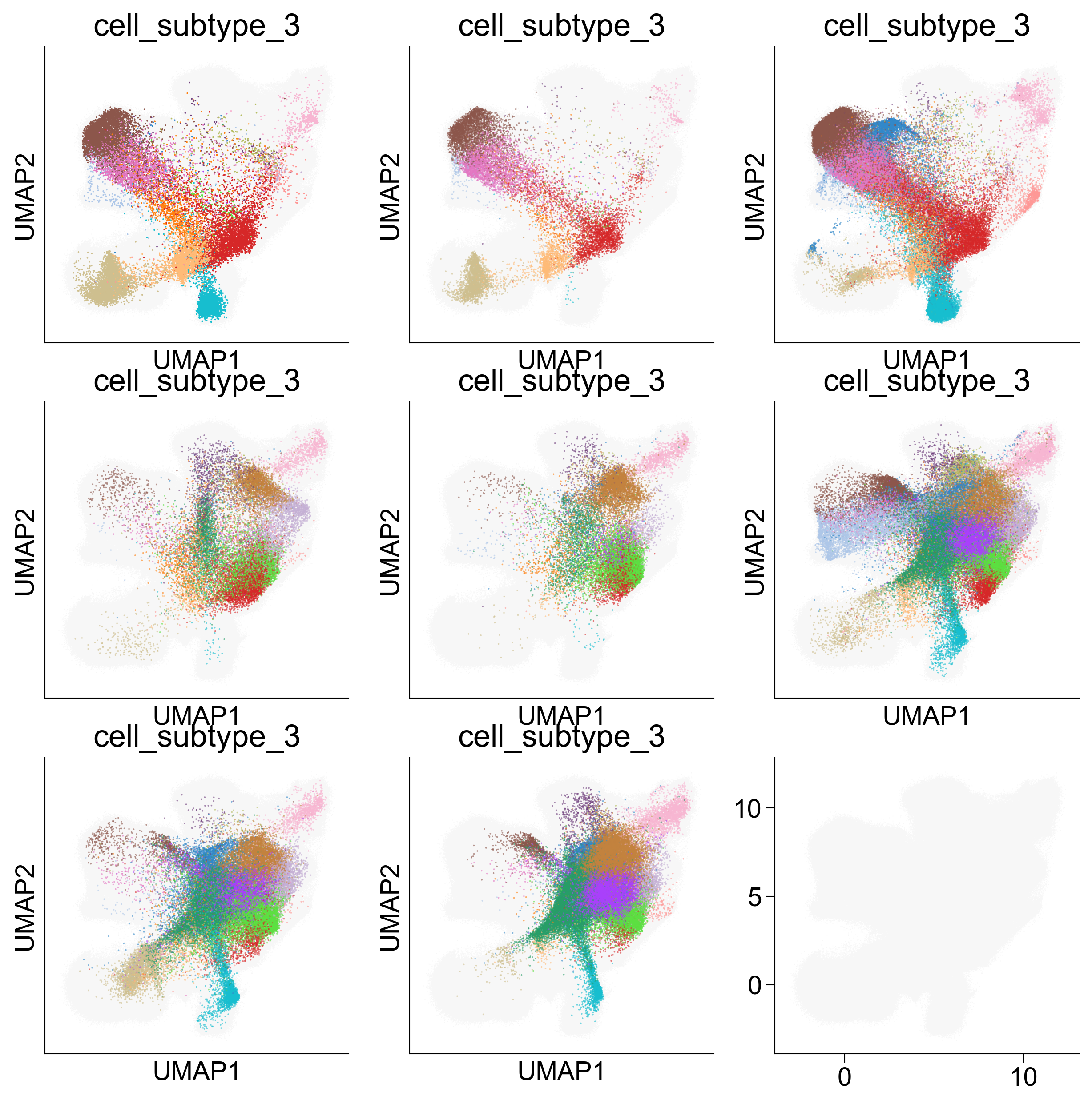
<Figure size 1800x1200 with 0 Axes>
Figure 5D¶
fraction, _ = plot_a_by_b(
adata_cd8[
np.array(adata_cd8.obs["disease_meta_type"] == "Solid tumor")
& np.array(adata_cd8.obs["tissue_meta_type"] == "TIL")
],
a="cell_subtype_3",
b="disease",
)
plt.close()
fraction.index = list(fraction.iloc[:, 0])
fraction = fraction.iloc[:, 1:]
sns.clustermap(
fraction.T.loc[
[
"Tn",
"Tcm",
"Early Tcm/Tem",
"GZMK+ Tem",
"GNLY+ Temra",
"CMC1+ Temra",
"ZNF683+ Teff",
"MAIT",
"ILTCK",
"ITGAE+ Trm",
"CREM+ Trm",
"ITGB2+ Trm",
"Tpex",
"GZMK+ Tex",
"ITGAE+ Tex",
"S100A11+ Tex",
"MACF1+ T",
"Cycling T",
]
],
cmap="Oranges",
xticklabels=1,
yticklabels=1,
vmax=0.7,
row_cluster=False,
)
plt.savefig("./figures/reference_dataset_solid_tumor_TIL.pdf")
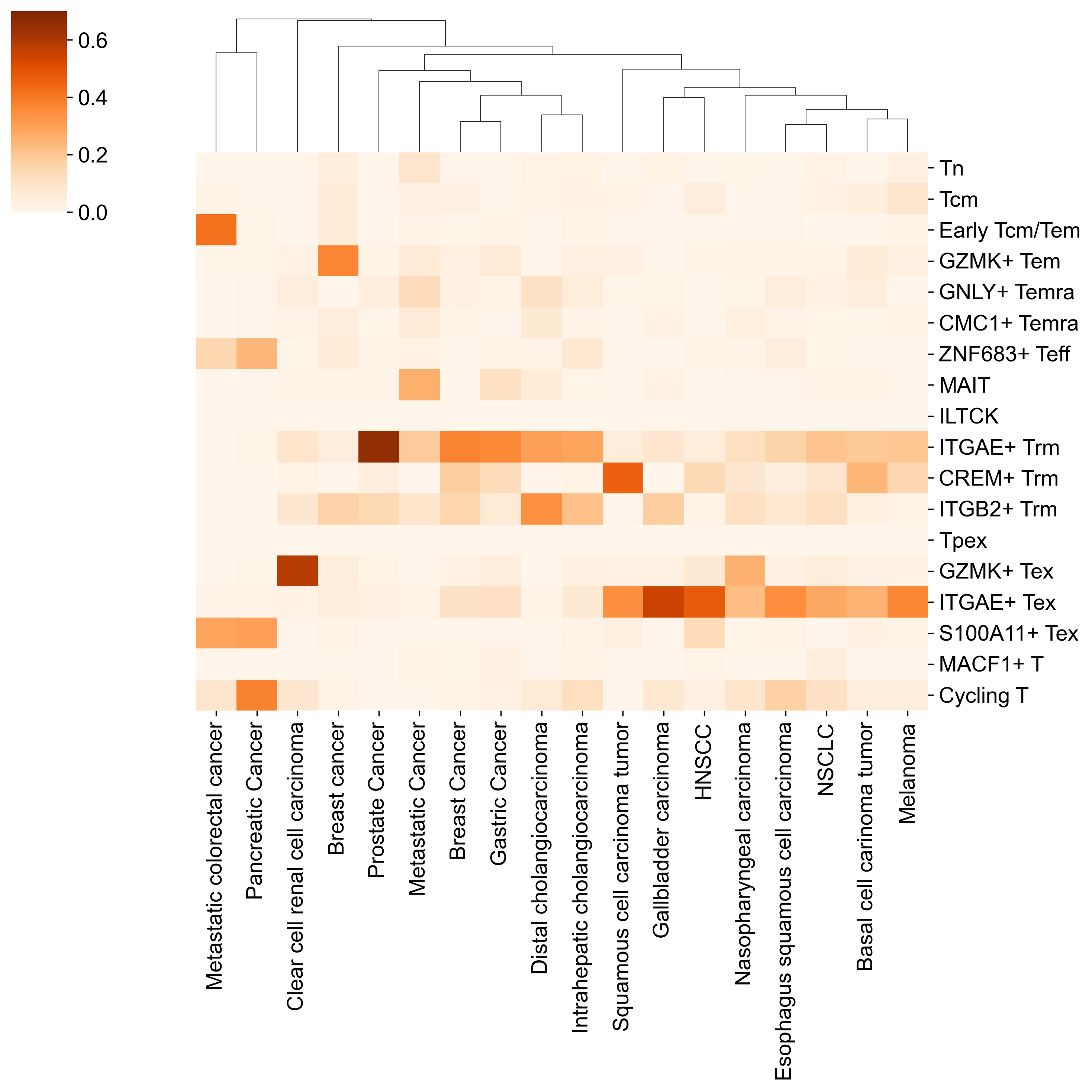
_merged_adata_for_transfer = merged_adata_for_transfer[
np.array(merged_adata_for_transfer.obs["tissue_type"] == "PBMC")
]
_merged_adata_for_transfer.obs["disease_treatment"] = list(
map(
lambda x: str(x[0]) + "-" + "Pre"
if "naive" in x[1]
else str(x[0]) + "-" + "Post",
zip(
_merged_adata_for_transfer.obs["disease_type"],
_merged_adata_for_transfer.obs["treatment"],
),
)
)
# sc.pl.umap(_merged_adata_for_transfer, color='cell_subtype_3', palette=subtype_color)
fraction, _ = plot_a_by_b(
_merged_adata_for_transfer, a="cell_subtype_3", b="disease_treatment"
)
plt.close()
fraction.index = list(fraction.iloc[:, 0])
fraction = fraction.iloc[:, 1:]
sns.heatmap(
fraction.T.loc[
[
"Tn",
"Tcm",
"Early Tcm/Tem",
"GZMK+ Tem",
"GNLY+ Temra",
"CMC1+ Temra",
"ZNF683+ Teff",
"MAIT",
"ILTCK",
"ITGAE+ Trm",
"CREM+ Trm",
"ITGB2+ Trm",
"Tpex",
"GZMK+ Tex",
"ITGAE+ Tex",
"S100A11+ Tex",
"MACF1+ T",
"Cycling T",
]
],
cmap="Blues",
xticklabels=1,
yticklabels=1,
vmax=0.5,
)
plt.savefig("./figures/transfered_dataset_solid_tumor_pbmc.pdf")

Extended Data Fig 12¶
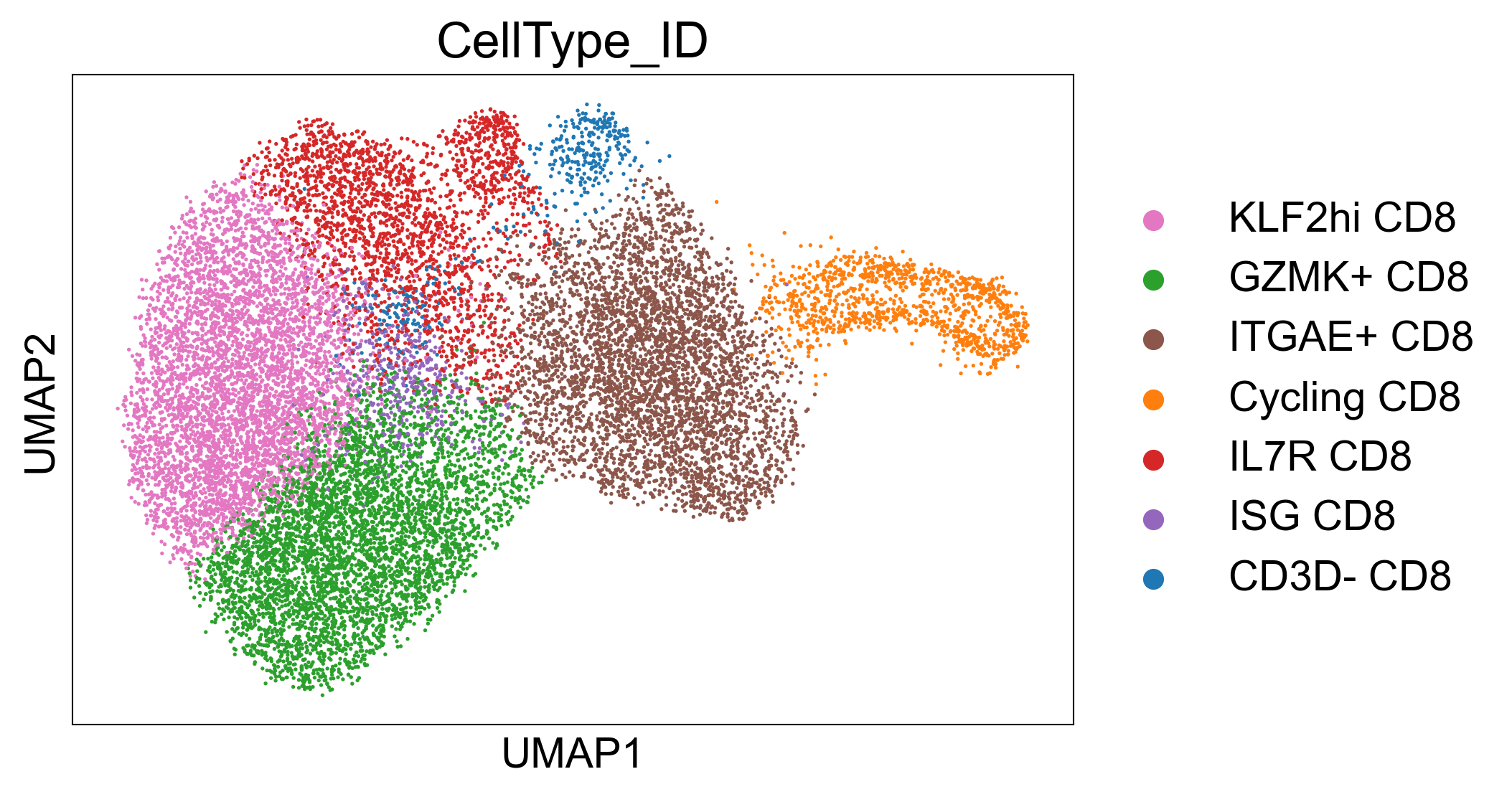
adata_luoma_til = sc.read_h5ad("./data/transfer_data/Luoma_2022_merged_TIL_CD8.h5ad")
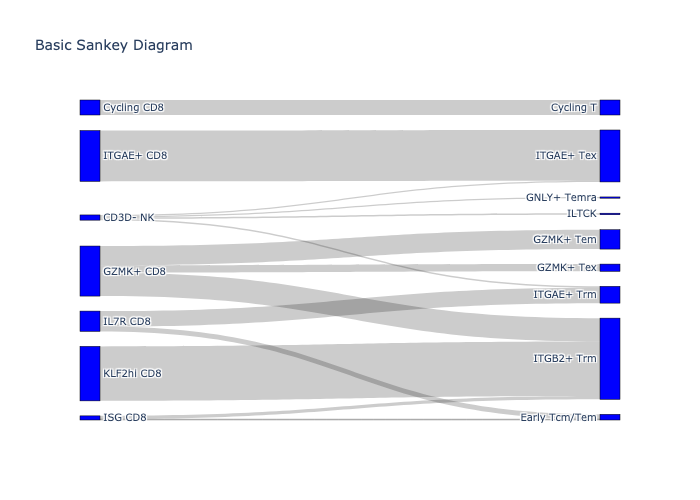
sc.pl.umap(adata_luoma_til, color='CellType_ID',palette=dict(zip([
'CD3D- CD8',
'Cycling CD8',
'GZMK+ CD8',
'IL7R CD8',
'ISG CD8',
'ITGAE+ CD8',
'KLF2hi CD8'], default_10)))
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(
count, fig = scatlasvae.tl.cell_type_alignment(
adata_luoma_til, obs_1="CellType_ID", obs_2="cell_subtype_3"
)
fig.show(renderer="png")
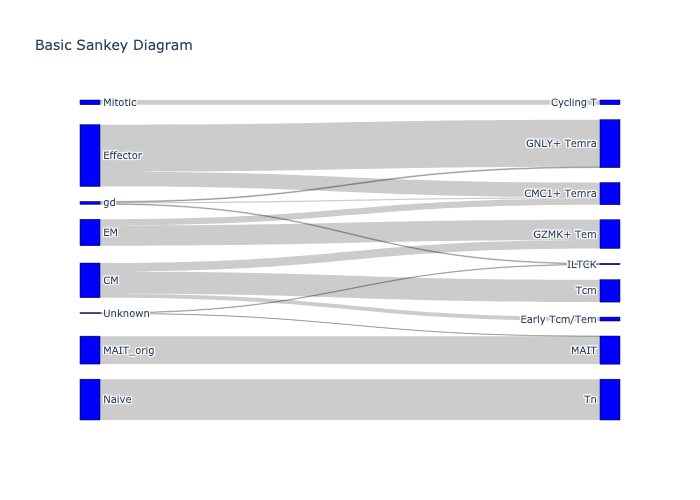
adata_luoma_2 = sc.read_h5ad("./data/transfer_data/Luoma_2022_merged_PBMC_CD8.h5ad")
sc.pl.umap(adata_luoma_2, color='CellType_ID', palette={
'CCR7+ CD8': '#3977BB',
'CD38+ CD8': '#E64540',
'DN T cells': '#009378',
'FGFBP2hi CD8': '#CB62A0',
'GZMBhi CD8': '#B8D391',
'GZMK+ CD8': '#EF9C00',
'IL7R+ CD8': '#69C5D9',
'KLRB1+ CD8': '#F6CF17',
'LTB+ CD8': '#D2D2CD'
})
/usr/local/lib/python3.9/site-packages/scanpy/plotting/_tools/scatterplots.py:394: UserWarning: No data for colormapping provided via 'c'. Parameters 'cmap' will be ignored
cax = scatter(

count, fig = scatlasvae.tl.cell_type_alignment(
adata_watson, obs_1="orig.ident", obs_2="cell_subtype_3"
)
fig.show(renderer="png")
Map Zheng et al., 2021 atlas to pan-disease CD8 T atlas Tex subset¶
Note
For training scAtlasVAE model you will need CUDA available.
Please see the PyTorch official website for installing GPU-enabled version of PyTorch.
adata_cd8_zheng_2021_tex_tpex.obs['cell_subtype_4'] = 'undefined'
adata_cd8_zheng_2021_tex_tpex.obs['cell_subtype_4'] = pd.Categorical(
list(adata_cd8_zheng_2021_tex_tpex.obs['cell_subtype_4']),
categories=adata_cd8_tex.obs['cell_subtype_4'].cat.categories
)
import torch
state_dict = torch.load(
"/rsch/Snowxue/2023-NM-Revision-GEX-HuARdb-Notebooks/20231210Tex.retrain_vae_for_transfer.model"
)
vae_model_transfer = scatlasvae.model.scAtlasVAE(
adata=adata_cd8_zheng_2021_tex_tpex,
pretrained_state_dict=state_dict['model_state_dict'],
**state_dict['model_config']
)
df = vae_model_transfer.predict_labels(return_pandas=True)
adata_cd8_zheng_2021_tex_tpex.obsm['X_retrain_GEX'] = vae_model_transfer.get_latent_embedding()
adata_cd8_zheng_2021_tex_tpex.obsm['X_retrain_umap'] = scatlasvae.tl.transfer_umap(
adata_cd8_tex.obsm['X_retrain_GEX'],
adata_cd8_tex.obsm['X_retrain_umap'],
adata_cd8_zheng_2021_tex_tpex.obsm['X_retrain_GEX'],
method='knn'
)['embedding']
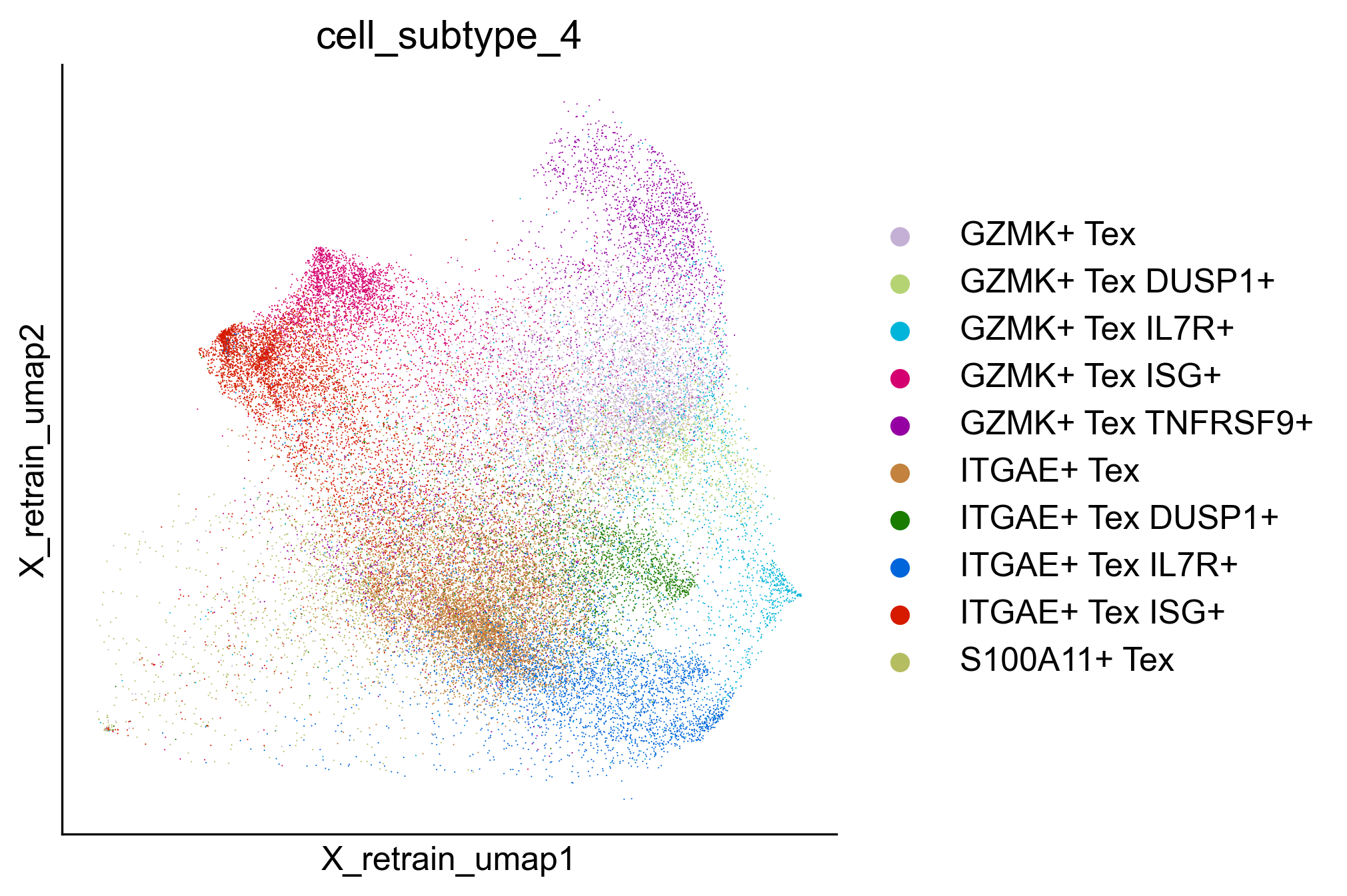
Extended Data Figure 10h¶
fig,ax=createFig(figsize=(5,5))
sc.pl.embedding(adata_cd8_zheng_2021_tex_tpex, basis='X_retrain_umap', color='cell_subtype_4', cmap='Reds', layer='normalized', s=1, palette={
'GZMK+ Tex':'#C5B0D5',
'GZMK+ Tex DUSP1+': '#B6D373',
'GZMK+ Tex IL7R+': '#00B4DA',
'GZMK+ Tex ISG+': '#D5006F',
'GZMK+ Tex TNFRSF9+': '#9500A3',
'ITGAE+ Tex':'#C3823E',
'ITGAE+ Tex DUSP1+': '#1A7C00',
'ITGAE+ Tex IL7R+': '#0065DA',
'ITGAE+ Tex ISG+': '#D51A00',
'S100A11+ Tex':'#b5bd61',
}, ax=ax)
Extended Data Figure 10j-m¶
adata_cd8_zheng_2021_tex_tpex.obs["disease_type"] = list(
map(lambda x: x.split(".")[0], adata_cd8_zheng_2021_tex_tpex.obs.index)
)
adata_cd8_zheng_2021_tex_tpex.obs["disease_type"] = list(
map(
lambda x: x[0] + "-" + x[1],
zip(
adata_cd8_zheng_2021_tex_tpex.obs["disease_type"],
adata_cd8_zheng_2021_tex_tpex.obs["study_name"],
),
)
)
obs = adata_cd8_tex.obs
obs = obs[
obs["study_name"].isin(
list(
map(
lambda z: z[0],
filter(lambda x: x[1] > 10, Counter(obs["study_name"]).items()),
)
)
)
]
obs = obs[
obs["sample_name"].isin(
list(
map(
lambda z: z[0],
filter(lambda x: x[1] > 10, Counter(obs["sample_name"]).items()),
)
)
)
]
sample_information = pd.read_csv(
"/Users/snow/Desktop/sample_information.csv", index_col=0
)
obs["tissue_type"] = list(sample_information.loc[obs["sample_name"], "Tissue Metatype"])
obs = obs[obs["tissue_type"] == "TIL"]
obs["disease_type"] = list(sample_information.loc[obs["sample_name"], "Disease"])
obs["disease_meta_type"] = list(
sample_information.loc[obs["sample_name"], "Disease Metatype"]
)
obs["treatment_status"] = list(
sample_information.loc[obs["sample_name"], "Treatment Status"]
)
obs = obs[obs["disease_meta_type"] == "Solid tumor"]
obs["disease_type"] = (
obs["disease_type"]
.replace("Melanoma", "Skin cancer")
.replace("Basal cell carinoma tumor", "Skin cancer")
.replace("Squamous cell carcinoma tumor", "Skin cancer")
)
obs["disease_type"] = list(
map(lambda x: x[1] + "-huARdb", zip(obs["study_name"], obs["disease_type"]))
)
sample_information_zheng_2021 = pd.read_csv(
"PanCancerdata.expression/expression/CD8/integration/int.CD8.S35.meta.tb.csv",
index_col=0,
)
sample_information_zheng_2021.index = list(sample_information_zheng_2021.iloc[:, 0])
adata_cd8_zheng_2021_tex_tpex.obs["tissue_type"] = sample_information_zheng_2021.loc[
adata_cd8_zheng_2021_tex_tpex.obs.index
]["loc"]
adata_cd8_zheng_2021_tex_tpex = adata_cd8_zheng_2021_tex_tpex[
adata_cd8_zheng_2021_tex_tpex.obs["tissue_type"] == "T"
]
obs = pd.concat(
[
obs,
adata_cd8_zheng_2021_tex_tpex.obs[
~adata_cd8_zheng_2021_tex_tpex.obs["study_name"].isin(
["BCC.KathrynEYost2019", "SCC.KathrynEYost2019"]
)
],
]
)
disease_palette = dict(zip(np.unique(obs["disease_type"]), sc.pl.palettes.godsnot_102[26:]))
c = Counter(obs["sample_name"])
agg = obs.groupby(["disease_type", "sample_name", "cell_subtype_4"]).agg(
{"sample_name": lambda x: len(x) / c[list(x)[0]]}
)
agg.columns = ["count"]
agg = pandas_aggregation_to_wide(agg[~agg.iloc[:, 0].isna()])
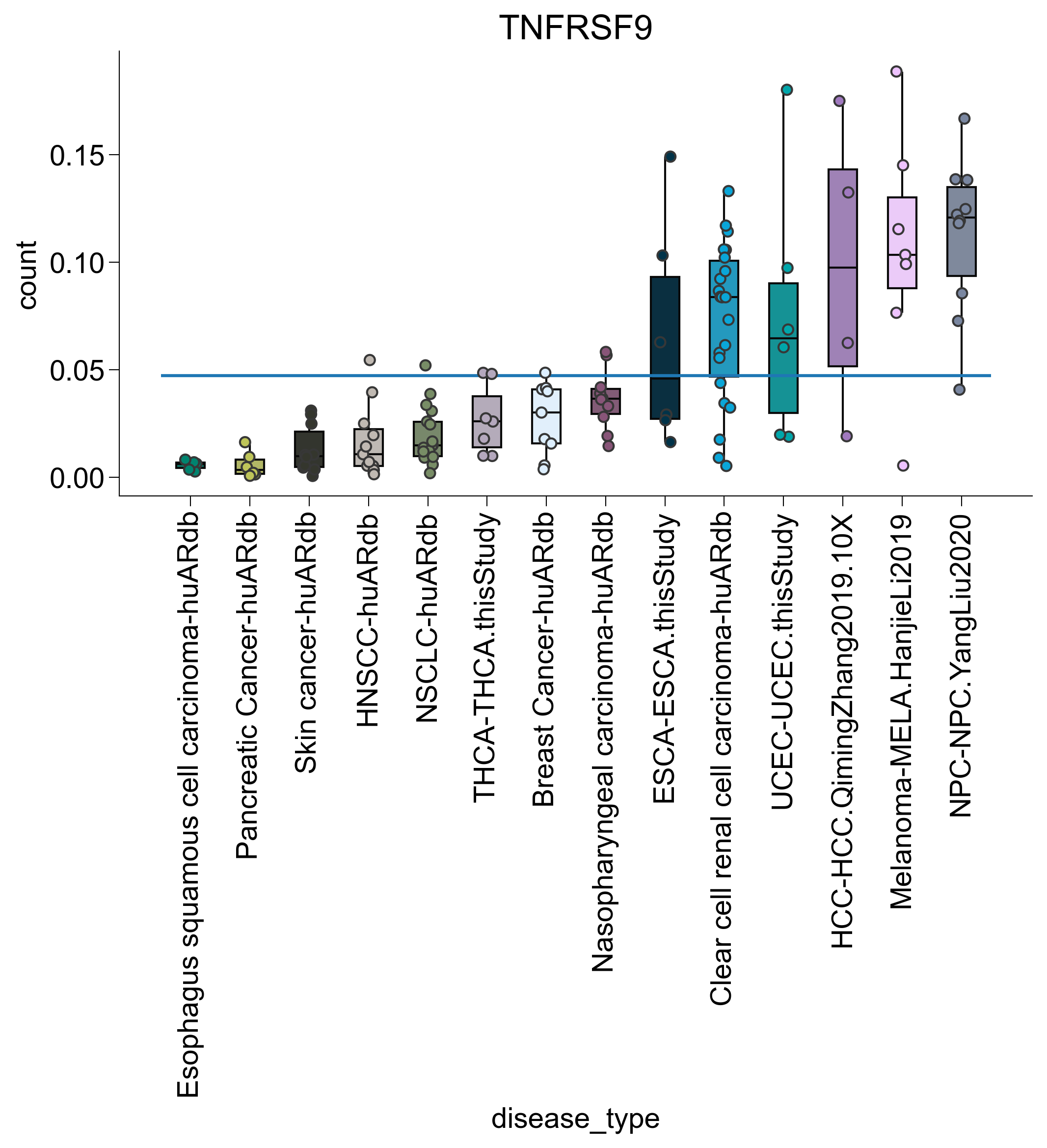
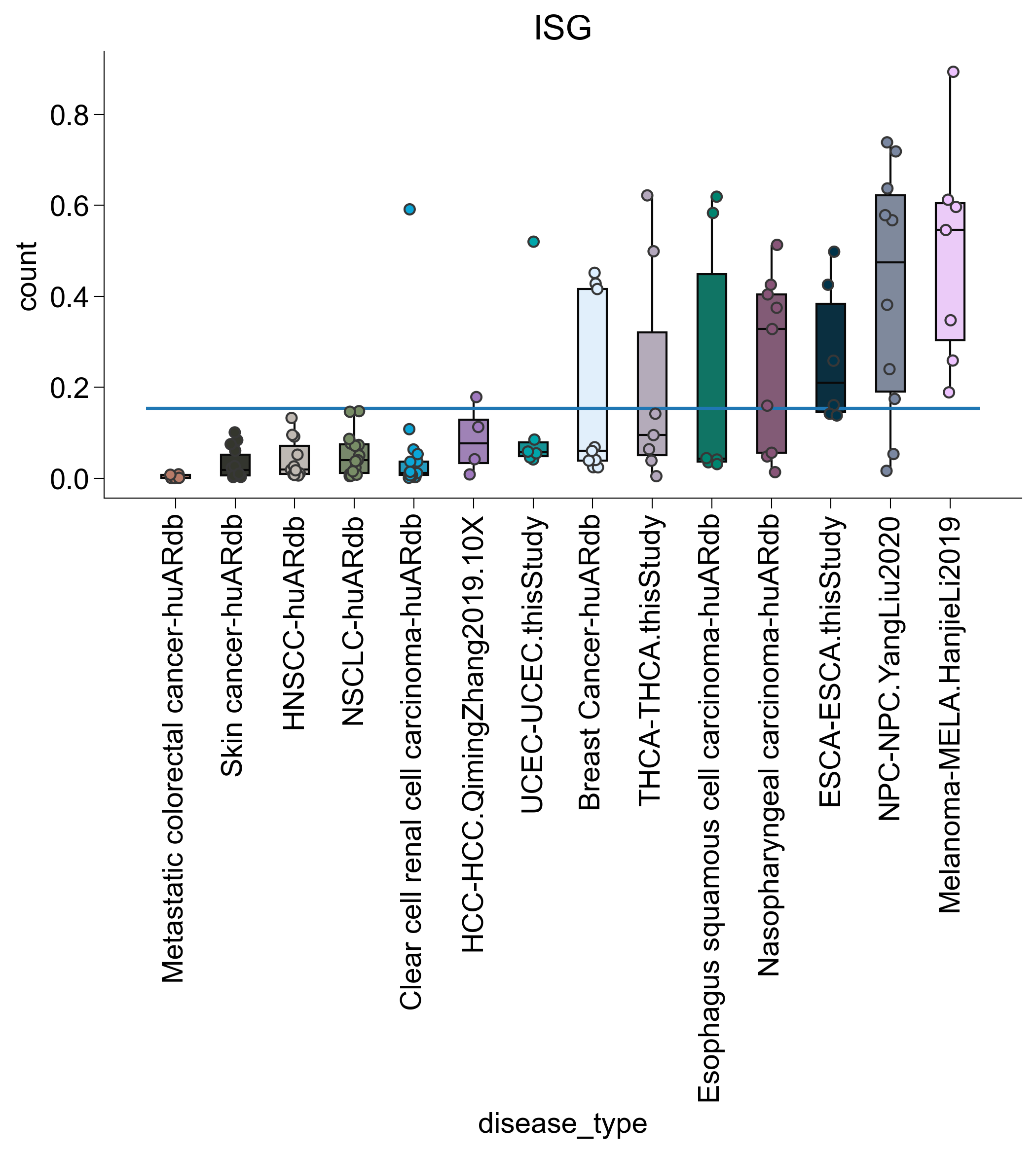
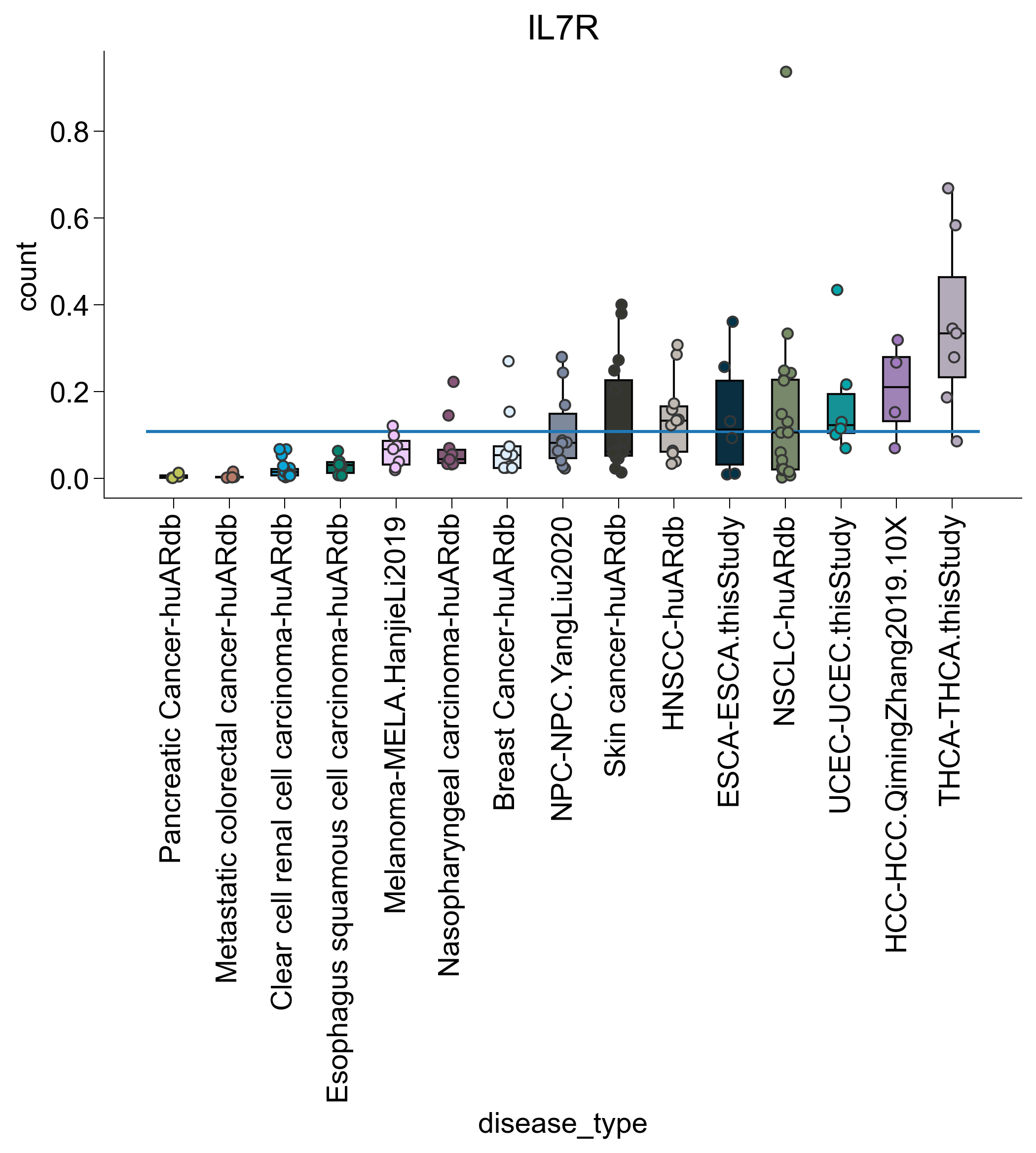
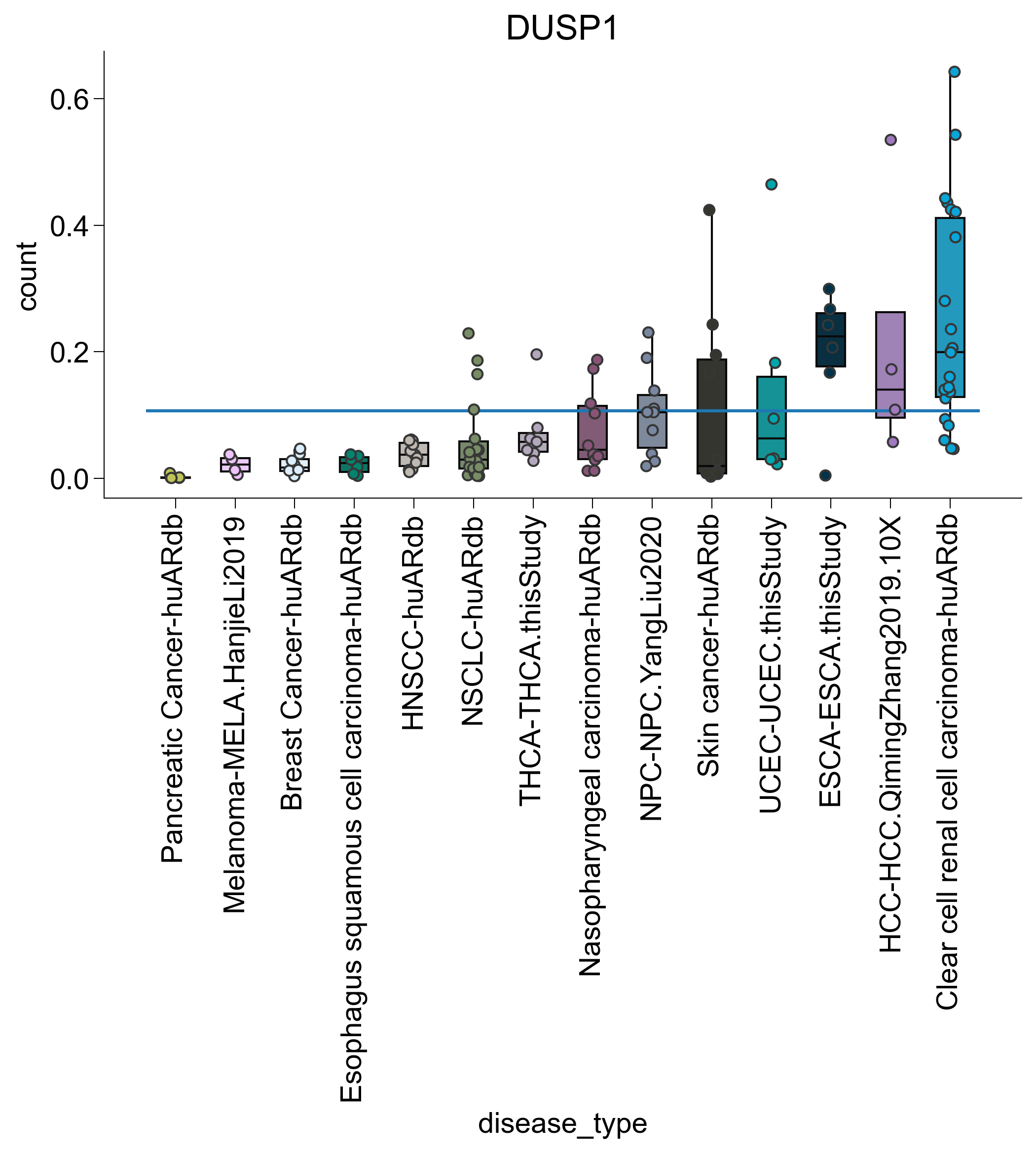
for subtype in ["TNFRSF9", "ISG", "IL7R", "DUSP1"]:
agg2 = pandas_aggregation_to_wide(
agg[list(map(lambda x: subtype in x, agg["cell_subtype_4"]))]
.groupby(["disease_type", "sample_name"])
.agg({"count": sum})
)
s = set(list(map(lambda z: z[0], filter(lambda x: x[1] > 100, c.items()))))
agg2 = agg2[list(map(lambda x: x in s, agg2["sample_name"]))]
s = set(
list(
map(
lambda x: x[0],
list(filter(lambda x: x[1] > 3, Counter(agg2["disease_type"]).items())),
)
)
)
agg2 = agg2[list(map(lambda x: x in s, agg2["disease_type"]))]
rank = dict(
zip(
agg2.groupby("disease_type")
.agg({"count": np.mean})
.sort_values("count")
.index,
range(len(np.unique(agg2["disease_type"]))),
)
)
agg2["disease_type_rank"] = list(map(lambda x: rank.get(x), agg2["disease_type"]))
agg2 = agg2.sort_values("disease_type_rank")
fig, ax = createFig()
sns.boxplot(
data=agg2,
x="disease_type",
y="count",
showfliers=False,
palette=disease_palette,
showcaps=False,
)
sns.stripplot(
data=agg2,
x="disease_type",
y="count",
dodge=False,
palette=disease_palette,
edgecolor="#373737",
linewidth=1,
)
ax.hlines(
xmin=ax.get_xbound()[0], xmax=ax.get_xbound()[1], y=np.mean(agg2["count"])
)
plt.xticks(rotation=90)
adjust_box_widths(fig, 0.6)
ax.set_title(subtype)